Interaction of R with databases on the example of Microsoft SQL Server and other DBMS
Since the lion's share of business information is stored in databases. In whatever programming language you write, you have to perform various actions with them.
In this article I will talk about two interfaces for working with databases in R. Most of the examples demonstrate working with Microsoft SQL Server, however, all code examples will work with other databases, such as: MySQL, PostgreSQL, SQLite, ClickHouse , Google BigQuery, etc.

Content
- Software required
- DBI package
- RODBC Package
- Conclusion
Software required
In order to repeat all the examples of working with the DBMS described in the article, you will need the following free software listed below:
- Language R ;
- RStudio development environment;
- Database Management System, to choose from:
3.1. Microsoft SQL Server
3.2. MySQL
3.3. PostgreSQL
DBI package
The DBI package is the most popular and convenient way to interact with databases in R.
DBI provides you with a set of functions with which you can manage databases. But to connect to databases, you need to install additional packages that are drivers for various database management systems (DBMS).
List of basic DBI functions
dbConnect- connection to the database;dbWriteTable- write a table to the database;dbReadTable- loading a table from a database;dbGetQuery- loading the result of query execution;dbSendQuery- sending a query to the database;dbFetch- extract elements from a result set;dbExecute- execution of requests to update / delete / insert data into tables;dbGetInfo- request information about the result of the request or connection;dbListFields- request for a list of table fields;dbListTables- query a list of database tables;dbExistsTable- check for the presence of a table in the database;dbRemoveTable- delete a table from the database;dbDisconnect- disconnect from database.
Database Connectivity
To interact with databases, you must first connect to them. Depending on the DBMS you plan to work with, you will need an additional package, below is a list of the most frequently used ones.
odbc- Driver for connecting via ODBC interface;RSQLite- Driver for SQLite;RMySQL/RMariaDB- Driver for MySQL and MariaDB;RPostgreSQL- Driver for PosrtgreSQL;bigrquery- Driver for Google BigQuery;RClickhouse/clickhouse- Driver for ClickHouse;RMSSQL- Driver for Microsoft SQL Server (MS SQL), at the time of writing this article is present only on GitHub .
The DBI package comes with the basic R package, but packages that are database drivers must be installed using the install.packages(" ") command install.packages(" ") .
To install packages from GitHub you will also need an additional package - devtools . For example, the RMSSQL package is not currently published in the main R package repository, use the following code to install it:
install.packages("devtools") devtools::install_github("bescoto/RMSSQL") An example of connecting to Microsoft SQL Server using odbc
Before using any package in an R session, it must first be connected using the library(" ") function library(" ") .
It was not without reason that I chose Microsoft SQL Server as the main DBMS, which will provide most of the examples in this article. The fact is that this is a fairly popular database, but at the same time it still does not have a driver for connecting from R published on CRAN.
But fortunately, SQL Server, like almost any other database, has an ODBC (English Open Database Connectivity) interface for connecting. There are a number of packages for connecting to the DBMS via the ODBC interface in R. First, we will look at connecting through the odbc package.
# odbc install.packages("odbc") # library(odbc) # MS SQL con <- dbConnect(drv = odbc(), Driver = "SQL Server", Server = "localhost", Database = "mybase", UID = "my_username", PWD = "my_password", Port = 1433) In the dbConnect() function, you need to pass the function, which is the driver for connecting to the DBMS ( odbc() ), as the first argument to drv . Such functions are usually called the same as the DBMS, and come with packages that are drivers for DBI .
Next, you need to list the connection parameters. To connect to MS SQL via ODBC, you must specify the following parameters:
- Driver - Name of the ODBC driver;
- Server - IP address of SQL server;
- Database - The name of the database to connect to;
- UID - Database username;
- PWD - Password;
- Port - The port to connect, with SQL Server, the default port is 1433.
An ODBC driver for connecting to Microsoft SQL Server is included with Windows, but it may have a different name. You can view the list of installed drivers in the ODBC Data Source Administrator. You can start the data source administrator in Windows 10 in the following way:
- 32-bit version:
%systemdrive%\Windows\SysWoW64\Odbcad32.exe - 64-bit version:
%systemdrive%\Windows\System32\Odbcad32.exe
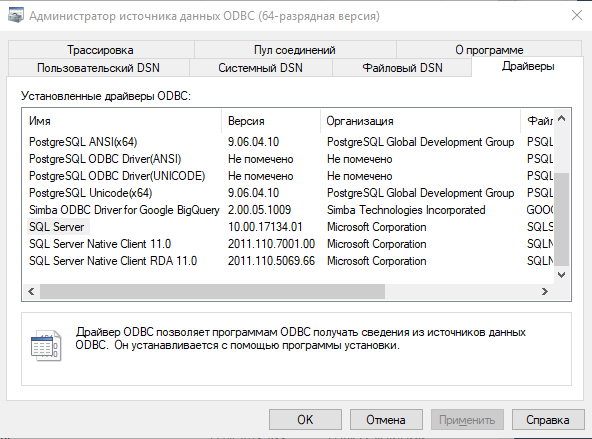
You can also get a list of all the drivers installed on your PC using the odbcListDrivers() function.
name attribute value <chr> <chr> <chr> 1 SQL Server APILevel 2 2 SQL Server ConnectFunctions YYY 3 SQL Server CPTimeout 60 4 SQL Server DriverODBCVer 03.50 5 SQL Server FileUsage 0 6 SQL Server SQLLevel 1 7 SQL Server UsageCount 1 8 MySQL ODBC 5.3 ANSI Driver UsageCount 1 9 MySQL ODBC 5.3 Unicode Driver UsageCount 1 10 Simba ODBC Driver for Google BigQuery Description Simba ODBC Driver for Google BigQuery2.0 # ... with 50 more rows You can download ODBC drivers for other DBMSs at the following links:
For various DBMSs, the name of the parameters for connection may be different, for example:
- PostgreSQL / MySQL / MariaDB - user, password, host, port, dbname;
- GoogleBigQuery - project, dataset;
- ClickHouse - user, password, db, port, host;
Using the ODBC data source administrator, you can run the wizard to create an ODBC data source. To do this, just open the administrator, go to the "Custom DSN" tab and click the "Add ..." button.

When creating a data source using an administrator, you give it a name, DSN (Data Source Name).

In the example above, we created a data source with the DSN "my_test_source". Now we can use this source to connect to Microsoft SQL Server, and not specify other connection parameters in the code.
# DSN con <- dbConnect(odbc(), DSN = "my_test_source", UID = "my_username", PWD = "my_password") You can view the names of all ODBC data sources created on your PC using the odbcListDataSources() function.
name description <chr> <chr> 1 BQ Simba ODBC Driver for Google BigQuery 2 BQ_main Simba ODBC Driver for Google BigQuery 3 BQ ODBC Simba ODBC Driver for Google BigQuery 4 OLX Simba ODBC Driver for Google BigQuery 5 Multicharts Simba ODBC Driver for Google BigQuery 6 PostgreSQL35W PostgreSQL Unicode(x64) 7 hillel_bq Simba ODBC Driver for Google BigQuery 8 blog_bq Simba ODBC Driver for Google BigQuery 9 MyClientMSSQL SQL Server 10 local_mssql SQL Server 11 MSSQL_localhost SQL Server 12 my_test_source SQL Server 13 Google BigQuery Simba ODBC Driver for Google BigQuery An example of connecting to Microsoft SQL Server using the RMSSQL package
RMSSQL not published on CRAN, so you can install it from GitHub using the devtools package.
install.packages("devtools") devtools::install_github("bescoto/RMSSQL") # library(RJDBC) library(RMSSQL) library(DBI) # RMSSQL con <- dbConnect(MSSQLServer(), host = 'localhost', user = 'my_username', password = 'my_password', dbname = "mybase") In most cases, using the DBI package to work with databases, you will connect in this way. Those. install one of the required driver packages, passing as the value of the drv argument to the dbConnect function, the function is the driver for connecting to the DBMS you need.
# MySQL library(RMySQL) con <- dbConnect(MySQL(), host = 'localhost', user = 'my_username', password = 'my_password', dbname = "mybase", host = "localhost") # PostrgeSQL library(RPostgreSQL) con <- dbConnect(PostgreSQL(), host = 'localhost', user = 'my_username', password = 'my_password', dbname = "mybase", host = "localhost") # PostrgeSQL library(RPostgreSQL) con <- dbConnect(PostgreSQL(), host = 'localhost', user = 'my_username', password = 'my_password', dbname = "mybase", host = "localhost") # SQLite library(RSQLite) # connection or create base con <- dbConnect(drv = SQLite(), "localhost.db") # Google BigQuery library(bigrquery) con <- dbConnect(drv = bigquery(), project = "my_proj_id", dataset = "dataset_name") Creating tables and writing to the database
Writing to the database is performed by the dbWriteTable() function.
Arguments to the dbWriteTable() function:
Required arguments are in bold, italics are optional
- conn - DBMS connection object created using the
dbConnectfunction; - name - the name of the table in the DBMS into which the data will be written;
- value - a table (an object of the data.frame / data.table / tibble_frame class) in R, the data from which will be written to the DBMS;
- row.names - Adds a row_names column with row numbers to the table, defaults to FALSE .
- overwrite - Overwrite the table if the table with the name specified in the name argument is already present in the DBMS; the default value is FALSE ;
- append - Append data if a table with the name specified in the name argument is already present in the DBMS; the default value is FALSE ;
- field.types - Accepts a named vector, and sets the data type in each field when writing to the DBMS, defaults to NULL ;
- temporary - Allows you to create temporary tables in the DBMS that will be available until the connection with the database is disconnected; the default value is FALSE .
# library(odbc) # DSN con <- dbConnect(odbc(), DSN = "my_test_source", UID = "my_username", PWD = "my_password") # iris, R iris dbWriteTable(conn = con, name = "iris", value = iris) # dbDisconnect(con) To view tables in the database, use the dbListTables() function, to delete tables dbRemoveTable()
# library(odbc) # con <- dbConnect(odbc(), DSN = "my_test_source", UID = "my_username", PWD = "my_password") # dbListTables(con) # iris dbRemoveTable(con, "iris") # dbDisconnect(con) Reading data from a DBMS
Using DBI you can query either entire tables or the result of executing your SQL query. The functions dbReadTable() and dbGetQuery() are used to perform these operations.
# library(odbc) # con <- dbConnect(odbc(), DSN = "my_test_source", UID = "my_username", PWD = "my_password") # iris iris dbiris <- dbReadTable(con, "iris") # dbDisconnect(con) # library(odbc) # con <- dbConnect(odbc(), DSN = "my_test_source", UID = "my_username", PWD = "my_password") # setosa <- dbGetQuery(con, "SELECT * FROM iris WHERE Species = 'setosa'") # dbDisconnect(con) Data manipulation in DBMS (DML)
The dbGetQuery() function dbGetQuery() above is used exclusively for querying data samples (SELECT).
For data manipulation operations, such as UPDATE, INSERT, DELETE, the dbExecute() function exists in DBI .
# library(odbc) # con <- dbConnect(odbc(), DSN = "my_test_source", UID = "my_username", PWD = "my_password") # (INSERT) dbExecute(con, "INSERT INTO iris (row_names, [Sepal.Length], [Sepal.Width], [Petal.Length], [Petal.Width], [Species]) VALUES (51, 5.0, 3.3, 1.7, 0.3, 'new_values')") # (UPDATE) dbExecute(con, "UPDATE iris SET [Species] = 'old_value' WHERE row_names = 51") # (DELETE) dbExecute(con, "DELETE FROM iris WHERE row_names = 51") # dbDisconnect(con) Transactions in the DBMS
A transaction is a sequential read and write operation. The end of a transaction can be either saving changes (commit, commit) or canceling changes (rollback, rollback). In relation to the database, a transaction is a series of queries, which are treated as a single query.
Quote from the article "Transactions and the mechanisms of their control"
A transaction encapsulates several SQL statements in an elementary unit. In DBI start of a transaction is initiated with dbBegin() and then either confirmed with dbCommit() or canceled with dbRollback() . In any case, the DBMS guarantees that: either all or none of the statements will be applied to the data.
For example, during the transaction, let's add 51 rows to the iris table, then change the Sepal.Width value to 5 lines, and delete 43 lines from the table.
# library(odbc) # con <- dbConnect(odbc(), DSN = "my_test_source", UID = "my_username", PWD = "my_password") # dbGetQuery(con, "SELECT * FROM iris WHERE row_names IN (5, 43, 51)") # row_names Sepal.Length Sepal.Width Petal.Length Petal.Width Species # 1 5 5.0 3.6 1.4 0.2 setosa # 2 43 4.4 3.2 1.3 0.2 setosa # 3 51 7.0 3.2 4.7 1.4 versicolor # dbBegin(con) # dbExecute(con, "INSERT INTO iris (row_names, [Sepal.Length], [Sepal.Width], [Petal.Length], [Petal.Width], [Species]) VALUES (51, 5.0, 3.3, 1.7, 0.3, 'new_values')") # dbExecute(con, "UPDATE iris SET [Sepal.Width] = 8 WHERE row_names = 5") # 43 dbExecute(con, "DELETE FROM iris WHERE row_names = 43") # dbCommit(con) # dbGetQuery(con, "SELECT * FROM iris WHERE row_names IN (5, 43, 51)") # row_names Sepal.Length Sepal.Width Petal.Length Petal.Width Species # 1 5 5 8.0 1.4 0.2 setosa # 2 51 7 3.2 4.7 1.4 versicolor # 3 51 5 3.3 1.7 0.3 new_values # library(odbc) # con <- dbConnect(odbc(), DSN = "my_test_source", UID = "my_username", PWD = "my_password") # dbGetQuery(con, "SELECT * FROM iris WHERE row_names IN (5, 43, 51)") # row_names Sepal.Length Sepal.Width Petal.Length Petal.Width Species # 1 5 5.0 3.6 1.4 0.2 setosa # 2 43 4.4 3.2 1.3 0.2 setosa # 3 51 7.0 3.2 4.7 1.4 versicolor # dbBegin(con) # dbExecute(con, "INSERT INTO iris (row_names, [Sepal.Length], [Sepal.Width], [Petal.Length], [Petal.Width], [Species]) VALUES (51, 5.0, 3.3, 1.7, 0.3, 'new_values')") # dbExecute(con, "UPDATE iris SET [Sepal.Width] = 8 WHERE row_names = 5") # 43 dbExecute(con, "DELETE FROM iris WHERE row_names = 43") # dbRollback(con) # dbGetQuery(con, "SELECT * FROM iris WHERE row_names IN (5, 43, 51)") # row_names Sepal.Length Sepal.Width Petal.Length Petal.Width Species # 1 5 5.0 3.6 1.4 0.2 setosa # 2 43 4.4 3.2 1.3 0.2 setosa # 3 51 7.0 3.2 4.7 1.4 versicolor RODBC Package
The RODBC package provides a stand-alone interface for connecting and working with a DBMS through an ODBC interface.
RODBC not DBI compatible, i.e. you cannot use the connection object created using RODBC in the functions provided by the DBI package.
Key features of the RODBC package
odbcConnect- Connection to the DBMS via DSN;odbcDriverConnect- Connection to the database through the connection string;sqlQuery- Sending a query to the DBMS, and getting the result of its execution. It supports any type of query: SELECT, UPDATE, INSERT, DELETE.sqlFetch- Get the whole table from the DBMS;sqlTables- Get a list of tables in the database.sqlSave- Create a new table in the database, or add new data to an existing table;sqlUpdate- Updating data in a table that already exists in the DBMS;sqlDrop- Delete a table in a DBMS;odbcClose- End a connection to a DBMS.
An example of working with RODBC
From my point of view, RODBC less functional than DBI , but it has all the necessary functions for working with a DBMS.
# library(RODBC) # con_string <- odbcDriverConnect(connection = "Driver=SQL Server;Server=localhost;Database=mybase;UID=my_username;PWD=my_password;Port=1433") # DSN con_dsn <- odbcConnect(dsn = "my_test_source", uid = "my_username", pwd = "my_password") # sqlSave(con_dsn, dat = iris, tablename = "iris") # iris sqlSave(con_dsn, dat = iris, tablename = "iris", append = TRUE) # 4 sqlFetch(con_dsn, "iris", rownames = FALSE, max = 4) # R iris[1, 5] <- "virginica" # sqlUpdate(con_dsn, dat = iris, tablename = "iris") # 4 sqlFetch(con_dsn, "iris", rownames = FALSE, max = 4) # sqlDrop(con_dsn, sqtable = "iris") # odbcCloseAll() Transactional
By default, transactionality in RODBC turned off. Transaction management is carried out by two functions.
odbcSetAutoCommit- Switch between the normal and transactional DBMS operation modes;odbcEndTran- Confirm or cancel a transaction.
Transactional mode is odbcSetAutoCommit and disabled by the odbcSetAutoCommit function using the autoCommit argument.
# library(RODBC) # DSN con_dsn <- odbcConnect(dsn = "my_test_source", uid = "my_username", pwd = "my_password") # sqlSave(con_dsn, dat = iris, tablename = "iris") # odbcSetAutoCommit(con_dsn, autoCommit = FALSE) # 4 sqlFetch(con_dsn, "iris", rownames = FALSE, max = 4) # rownames SepalLength SepalWidth PetalLength PetalWidth Species # 1 1 5.1 3.5 1.4 0.2 setosa # 2 2 4.9 3.0 1.4 0.2 setosa # 3 3 4.7 3.2 1.3 0.2 setosa # 4 4 4.6 3.1 1.5 0.2 setosa # R iris[1, 5] <- "virginica" # sqlUpdate(con_dsn, dat = iris, tablename = "iris") # 4 sqlFetch(con_dsn, "iris", rownames = FALSE, max = 4) # rownames SepalLength SepalWidth PetalLength PetalWidth Species # 1 1 5.1 3.5 1.4 0.2 virginica # 2 2 4.9 3.0 1.4 0.2 setosa # 3 3 4.7 3.2 1.3 0.2 setosa # 4 4 4.6 3.1 1.5 0.2 setosa # odbcEndTran(con_dsn, commit = FALSE) # 4 sqlFetch(con_dsn, "iris", rownames = FALSE, max = 4) # rownames SepalLength SepalWidth PetalLength PetalWidth Species # 1 1 5.1 3.5 1.4 0.2 setosa # 2 2 4.9 3.0 1.4 0.2 setosa # 3 3 4.7 3.2 1.3 0.2 setosa # 4 4 4.6 3.1 1.5 0.2 setosa # sqlUpdate(con_dsn, dat = iris, tablename = "iris") # odbcEndTran(con_dsn, commit = TRUE) # 4 sqlFetch(con_dsn, "iris", rownames = FALSE, max = 4) # rownames SepalLength SepalWidth PetalLength PetalWidth Species # 1 1 5.1 3.5 1.4 0.2 virginica # 2 2 4.9 3.0 1.4 0.2 setosa # 3 3 4.7 3.2 1.3 0.2 setosa # 4 4 4.6 3.1 1.5 0.2 setosa # odbcClose(con_dsn) Conclusion
The two methods of working with databases in the R, DBI and RODBC languages described in the article are quite universal and will work with almost any DBMS.
The only difference in operation between different DBMSs is the connection process. For most popular DBMSs, there are separate packages that are drivers. For the rest of the DBMS, it is necessary to configure the connection through the ODBC interface using odbc or RODBC . All other manipulations, regardless of which DBMS you have chosen, will be unchanged. An exception is sending SQL queries, depending on the SQL dialect supported by the DBMS you are working with.
')
Source: https://habr.com/ru/post/461063/
All Articles