Tasks of the domestic sequencer (June, 2019)
Before reading this upgrade, it is advisable to familiarize yourself with the previously published source .
The very first and most popular Minion nanoporous sequencer (the minimum set in the UK is $ 1000, in Russia is 150 thousand rubles), developed by Oxford Nanopore Technologies (ONT), works on disposable cells, each of which costs $ 900 ( Russia - 135 thousand rubles.). Such cells allow for 2 ... 3 days to digitize DNA with a length of 10 ... 20 Gb. This is too little for sequencing the human genome (need> 100 Gb), but too much for all other clinical diagnostic purposes (<1 Gb is enough).
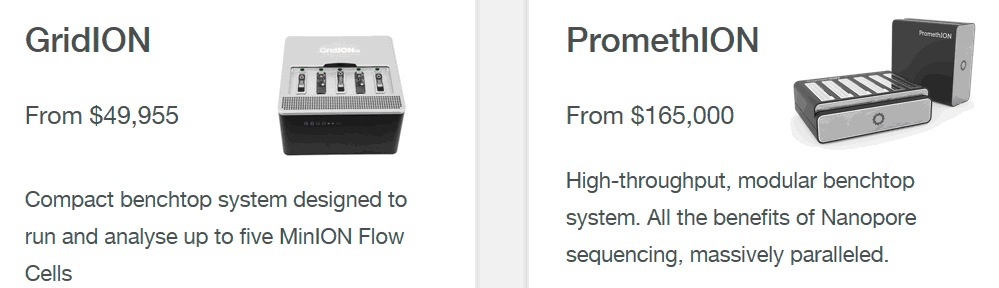
Recently, sales of more productive (and more expensive) models started - PromethION 24/48 ($ 165,000 / $ 285,000), aimed mainly at continuous genome-wide sequencing. More expensive are their more efficient consumable cells ($ 2,000 / piece,> 100 Gb). True, they can cost in bulk cheaper ($ 1,800,000 for 720 pieces, $ 625 / piece), but for the Russian budget such acquisitions (200 ... 250 million rubles per batch) are impermissible. And not only for the Russian. Therefore, the attention of visitors to the recent ONT conference (May 22-24) attracted a message about the start of sales of Flongle - an adapter insert to the MinION sequencer, which allows working with less efficient (~ 1 Gb) but relatively cheap ($ 90) disposable cells.

')
https://nanoporetech.com/products
The appearance of Flongle can cause an explosive growth in the use of NGS (next generation sequencing) in clinical practice and press off conventional PCR diagnostics, the market size of which in Russia exceeds one billion rubles, and is measured in the world by billions of dollars. But for the majority of Russian patients, NGS diagnostics will still remain an unaffordable luxury, since the cost of cells to Flongle in the domestic market will be above 10 thousand rubles. And if you add to it the cost of consumable reagents, transport costs and overhead costs, then the estimated cost of sequencing a single DNA / RNA sample will not fall below 20 thousand rubles.
Hence the need for the development of domestic nanoporous sequencers and their provision of cheap consumables and reagents.
It is possible to significantly reduce the cost of nanopor sequencing by reusing the Flongle cells and importing the consumables necessary for working with them. But first you need to get a test device that allows you to monitor the formation of bilayer lipid membranes (BLM) on 126 wells of the working cell, and assess the quantity and quality of nanopores embedded in these membranes. Such a device (“Nanorider”?) Should track changes in electrical conductivity (and / or impedance) of individual wells when BLM is formed on them, and record pico-ampere currents flowing through single ion channels.
The reader can be connected to a computer via USB 2.0 from standard elements: FPGA ($ 100 ... $ 200), current amplifiers (MAX9923FEUB - 200 ... 300 rubles), ADC (1 ... 2 thousand rubles), etc. ... Only a board with a non-standard contact pad (10x13) of type LGA (Land Grid Array) for docking with a cell from Flongle. Or with its simplified domestic analogue, containing a smaller number (16 ... 32) of sensory wells.
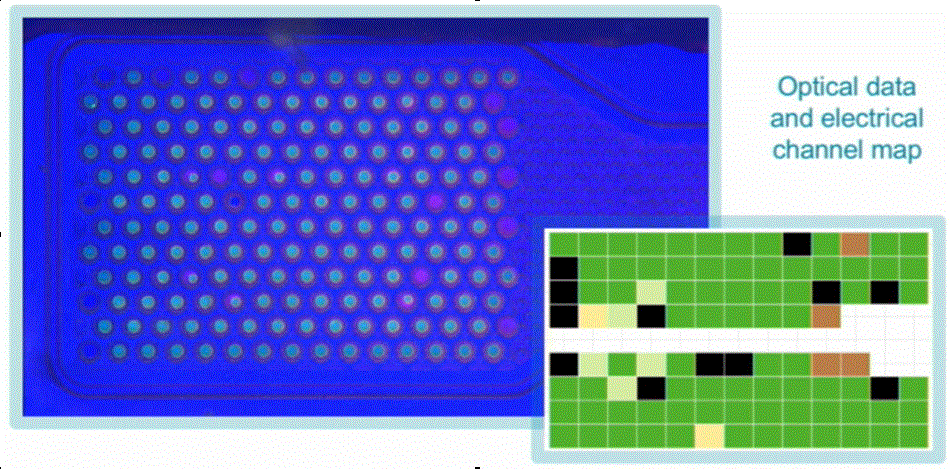
Flongle cell touch pad
Reducing the calculated cell performance by 4 ... 8 times (up to 10 ... 20 Mb) is not critical for solving many tasks (determining pathogens, HLA typing, paternity determination, personal identification, etc.), but it allows you to assemble a nanoporous sequencer on conventional amplifiers picoampere currents. And do without the use of special 512-channel chips, in the development of which the Oxfords invested tens of millions of dollars. True, each multichannel nanorider should contain more than a dozen current amplifiers, but the ADC can still be several times less than the proprietary combination of MinION + Flongle (> 200 thousand rubles). As for the poor performance of the nanorider, then for Russian users who do not have decent computers (doctors, biologists, biohackers, etc.), it may even be useful.
The Oxford-based 512-channel pico-amp current amplifiers are contained in each disposable cell to the MinION. Spent cells can be thrown away, but it is better to use the chips contained in them for the production of Flongle analogs characterized by increased productivity. If these chips are equipped with surface contacts, then they can work with contact cells containing 2048 (512x4) sensory wells each.
Developing cells with such LGAs will not be easy, but it must be borne in mind that the requirements for contacts are less stringent than those for processors and motherboards, for which one poor contact can destroy the entire system. For a sequencer, the inoperability of even half the contacts of a cell can be considered acceptable. The remaining half is enough to ensure performance at 5 ... 10 Gb.
The logical development of this idea will be the unification of several similar readers in one device. This will allow you to get an analogue of the GridION X5 sequencer, designed for simultaneous operation with five (or more) reusable cells of domestic production. Such a sequencer can be in demand in many clinical diagnostic laboratories.
https://store.nanoporetech.com/devices
PromethION disposable sequencer cells contain 9,000 sensor wells on chips. And the chips themselves are 3000-channel pico-amp current amplifiers. The flow of information cells issued by them from 24 (for PromethION 24) or from 48 (for PromethION 48) is so large that in its “raw” form it cannot be transferred to a regular external computer. Therefore, sequencers of this type are necessarily equipped with their own supercomputer. But if the sequencer will work with only one cell, then it can be connected to a regular gaming laptop with a Thunderbolt 3.0 port.
Alteration of free (ejected) chips from ONT to work with contact cells is not an insoluble problem. But sequencers based on the use of such chips can only work with ONT software. And the efforts that will have to be spent on reworking these programs can negate the benefits of using ready-made chips. And to develop such chips from scratch is too expensive.
This year, beta testing of the AXBIO (USA) nanoporous sequencer should begin. It uses chips with a million channels of current amplification, produced in Japan by the Israeli company Tower Semiconductor Ltd ... Expendable reagents and sales of these sequencers will be handled by the Chinese branch of the company AXBIO.

In the United States and the European Union, more than 7,000 patents are devoted to nanoporus sequencing, which creates unpredictable financial risks for local companies trying to engage in similar technologies. Apparently, this is the main reason for the high internationalization of this development. And allows Russian companies and laboratories to participate in the testing and improvement of the AXBIO technology without any restrictions, and also to produce (or assemble) such sequencers on its territory.
It is advisable to filter terabytes of raw data received by a nanoporous sequencer with megapixel chips (megarider). And transfer to the computer only the longest and high-quality reads. This will not only improve the quality of sequencing, but also reduce computer requirements, which is also important in mass production and widespread use of genomic sequencers. In addition, the excessive performance of mega-readers can reduce the duration of sequencing of the human genome from 2 ... 3 days (on ONT) to several hours.
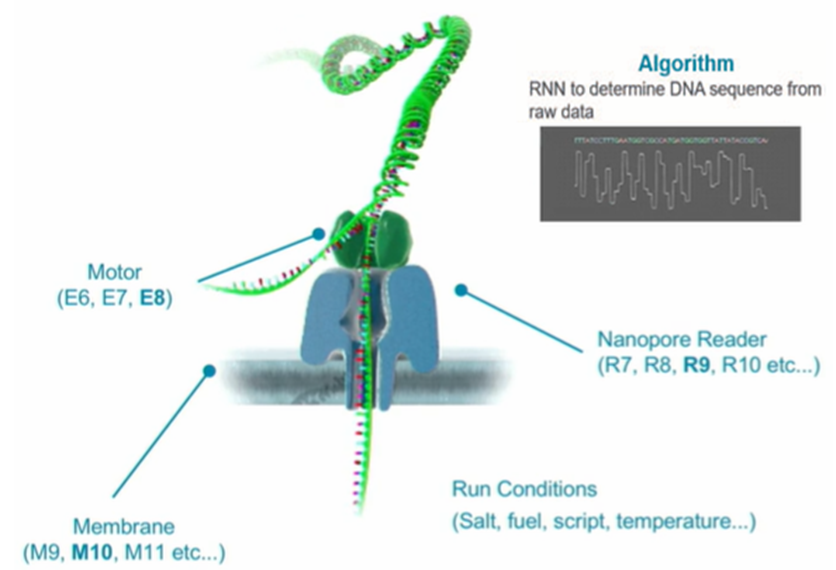
The success of ONT was facilitated by the organization of parallel conveyor optimization of the five basic elements of nanopore sequencing technology - a membrane, a nanopore, a motor (helicase), sequencing conditions, and decoding algorithms of read signals.
https://nanoporetech.com/resource-centre/videos/sub1000
The guaranteed shelf life of cells for nanoporous sequencing depends mainly on the stability of the bilayer lipid membrane. Now it is 6 weeks, although usually such membranes are destroyed within a few days. To increase their shelf life, hydrophobic polymer additives, surface-active block copolymers, cholesterol, antioxidants, cross-linking of fatty acids, etc. are used, and such modifications can interfere with the formation of ion channels. Therefore, to achieve stability BLM is very difficult.
This problem can be fundamentally solved by the formation of membranes in the working cells of the sequencer and the introduction of nanopores into them immediately before use. Independent preparation of cells for work will provide the possibility of their regeneration and repeated use. As for the shelf life, for dry cells it is practically unlimited, and the reagents necessary for their activation and regeneration can be stored for months.
The bottleneck in the technology of nanoprovod sequencing is at the same time the bottleneck of the nanopore, which determines the quality of the read information. Therefore, one of the main tasks of ONT was the search for membrane protein, which allows to obtain results of acceptable quality.
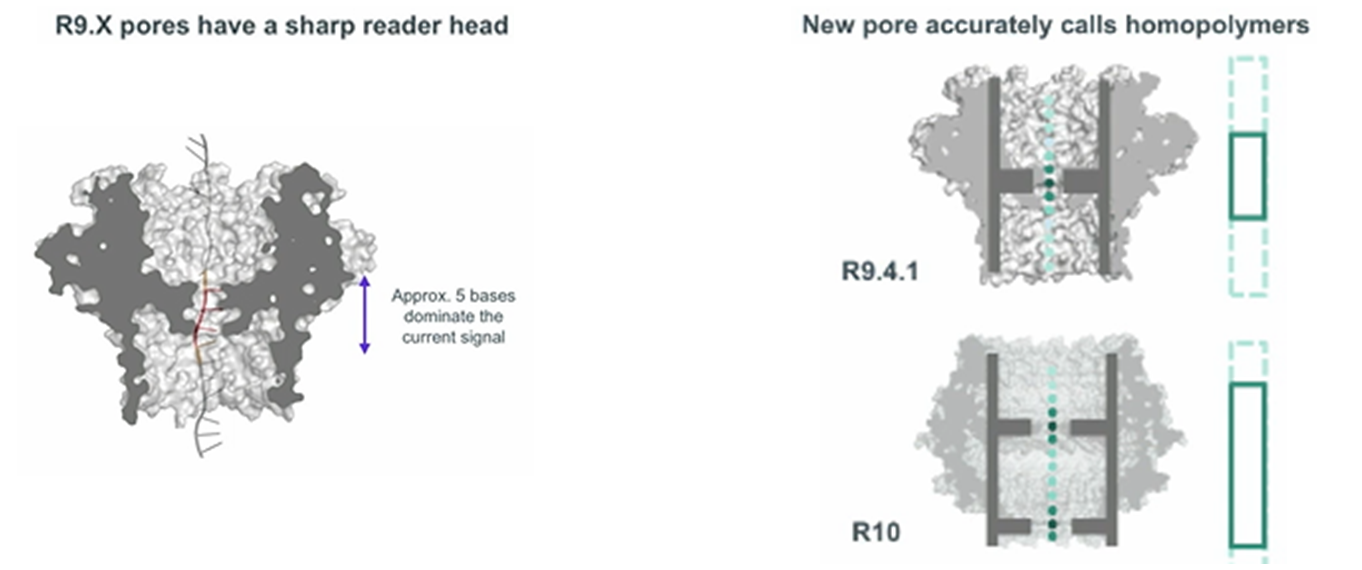
The first such protein was CsgG, an outer membrane protein found in many gram-negative bacteria. Its nine subunits form an ion channel in the membrane with wide entrance and exit vestibules, and one bottleneck. But it was possible to obtain sufficiently high-quality nanopores (R9, R9.4.1, R9.5.1) only after searching and analyzing hundreds of modified CsgG variants. This summer, cells with the next generation pores (R10 or R10b), characterized by increased accuracy of reading homopolymer repeats, should appear on sale.
https://nanoporetech.com/about-us/news/london-calling-clive-brown-and-team-plenary
The obligatory component of ONT's nanoporos sequencing technology is helicase, a molecular motor that unwinds double-stranded DNA and inhibits the promotion of single-stranded DNA through a nanopore. The maximum speed of DNA unwinding by such a motor now reaches 450 base pairs per second, although the speed of the electronic components of the MinION makes it possible to increase it to 1000 nucleotides per second. Theoretically, the acceleration of the work of helicase can double the performance of the MinION, but in practice this may lead to a deterioration in the quality of sequencing. Therefore, faster and more efficient helicases are unlikely to improve the performance of nanoporous sequencers such as MinION and PromethION.
The quality of sequencing depends on a variety of conditions - the magnitude of the voltage applied to the membrane, the electrical conductivity and the ionic composition of the reaction mixture, the amount and nature of impurities plugging the pores, the ability to clean the blocked pores by changing the polarity of the potential, the structure of the adapters used, etc., etc. Therefore, nanoporous sequencing requires careful optimization of all its parameters and compositions of working solutions (Run Conditions) that can adversely affect the quality of digitization of nucleotide followers awns. For example, it has recently been shown that reducing the concentration of ATP cleaved by helicase gradually reduces the accuracy of sequencing results, and now Oxfords are trying to eliminate this problem by introducing ATP regeneration system into the reaction mixture.
The fifth element of nanopor sequencing technology is artificial intelligence, built on recurrent neural networks (RNN) and on the use of some special algorithms and programs that improve the recognition of homogeneous repetitions (Flip-flop, Medaka). In July, the next release of the Guppi and MinKNOW programs should appear, in which the Oxfords promise to add the possibility of identifying methylated bases (5mC and 6mA). This is important not only for epigenomic studies, but also to improve the recognition of common bases.
The large volumes of information read by nanopor sequencer and the complexity of its processing impose increased requirements on the hardware of such software and hardware complexes. Therefore, not every computer is suitable for connecting MinION, GridION and PromethION put the latest generation of graphics accelerators, and Oxford Nanopore Technologies became one of the first buyers of a large batch of Jetson AGX Xavier (NVIDIA) modules designed for devices with artificial intelligence.
More recently, nanoporous technology was considered nothing more than a useful addition to more accurate fluorescent and semiconductor sequencing technologies. But a gradual increase in the accuracy of DNA reading, combined with a large length of readable sequences, allowed her, if not to get ahead of all competitors, then at least determine the main vector of the development of genomic sequencing technologies.

A lot of time may be needed to master all the elements of nanopor sequencing technology, but this is not the biggest problem. The main problem is the lack of targeted financing of such complex developments in Russia. True, the recently approved Federal Scientific and Technical Program for the Development of Genetic Technologies for 2019–2027 (section “Program Implementation Directions”) says that in the short term (3 ... 6 years) a prototype of the device will be developed for high-performance genomic sequencing. But to find this single line on 30 pages of text is not so easy. It is easier to agree with the Chinese on the purchase of AXBIO sequencers.
It is even easier to find craftsmen who can assemble the simplest nanorider “on their knees”. And then walk with an outstretched hand in search of investors who will agree to finance the development of domestic technology of nanoporous sequencing.
The very first and most popular Minion nanoporous sequencer (the minimum set in the UK is $ 1000, in Russia is 150 thousand rubles), developed by Oxford Nanopore Technologies (ONT), works on disposable cells, each of which costs $ 900 ( Russia - 135 thousand rubles.). Such cells allow for 2 ... 3 days to digitize DNA with a length of 10 ... 20 Gb. This is too little for sequencing the human genome (need> 100 Gb), but too much for all other clinical diagnostic purposes (<1 Gb is enough).
Recently, sales of more productive (and more expensive) models started - PromethION 24/48 ($ 165,000 / $ 285,000), aimed mainly at continuous genome-wide sequencing. More expensive are their more efficient consumable cells ($ 2,000 / piece,> 100 Gb). True, they can cost in bulk cheaper ($ 1,800,000 for 720 pieces, $ 625 / piece), but for the Russian budget such acquisitions (200 ... 250 million rubles per batch) are impermissible. And not only for the Russian. Therefore, the attention of visitors to the recent ONT conference (May 22-24) attracted a message about the start of sales of Flongle - an adapter insert to the MinION sequencer, which allows working with less efficient (~ 1 Gb) but relatively cheap ($ 90) disposable cells.

')
https://nanoporetech.com/products
The appearance of Flongle can cause an explosive growth in the use of NGS (next generation sequencing) in clinical practice and press off conventional PCR diagnostics, the market size of which in Russia exceeds one billion rubles, and is measured in the world by billions of dollars. But for the majority of Russian patients, NGS diagnostics will still remain an unaffordable luxury, since the cost of cells to Flongle in the domestic market will be above 10 thousand rubles. And if you add to it the cost of consumable reagents, transport costs and overhead costs, then the estimated cost of sequencing a single DNA / RNA sample will not fall below 20 thousand rubles.
Hence the need for the development of domestic nanoporous sequencers and their provision of cheap consumables and reagents.
Nanorider
It is possible to significantly reduce the cost of nanopor sequencing by reusing the Flongle cells and importing the consumables necessary for working with them. But first you need to get a test device that allows you to monitor the formation of bilayer lipid membranes (BLM) on 126 wells of the working cell, and assess the quantity and quality of nanopores embedded in these membranes. Such a device (“Nanorider”?) Should track changes in electrical conductivity (and / or impedance) of individual wells when BLM is formed on them, and record pico-ampere currents flowing through single ion channels.
The reader can be connected to a computer via USB 2.0 from standard elements: FPGA ($ 100 ... $ 200), current amplifiers (MAX9923FEUB - 200 ... 300 rubles), ADC (1 ... 2 thousand rubles), etc. ... Only a board with a non-standard contact pad (10x13) of type LGA (Land Grid Array) for docking with a cell from Flongle. Or with its simplified domestic analogue, containing a smaller number (16 ... 32) of sensory wells.

Flongle cell touch pad
Reducing the calculated cell performance by 4 ... 8 times (up to 10 ... 20 Mb) is not critical for solving many tasks (determining pathogens, HLA typing, paternity determination, personal identification, etc.), but it allows you to assemble a nanoporous sequencer on conventional amplifiers picoampere currents. And do without the use of special 512-channel chips, in the development of which the Oxfords invested tens of millions of dollars. True, each multichannel nanorider should contain more than a dozen current amplifiers, but the ADC can still be several times less than the proprietary combination of MinION + Flongle (> 200 thousand rubles). As for the poor performance of the nanorider, then for Russian users who do not have decent computers (doctors, biologists, biohackers, etc.), it may even be useful.
Kilorider
The Oxford-based 512-channel pico-amp current amplifiers are contained in each disposable cell to the MinION. Spent cells can be thrown away, but it is better to use the chips contained in them for the production of Flongle analogs characterized by increased productivity. If these chips are equipped with surface contacts, then they can work with contact cells containing 2048 (512x4) sensory wells each.
Developing cells with such LGAs will not be easy, but it must be borne in mind that the requirements for contacts are less stringent than those for processors and motherboards, for which one poor contact can destroy the entire system. For a sequencer, the inoperability of even half the contacts of a cell can be considered acceptable. The remaining half is enough to ensure performance at 5 ... 10 Gb.
The logical development of this idea will be the unification of several similar readers in one device. This will allow you to get an analogue of the GridION X5 sequencer, designed for simultaneous operation with five (or more) reusable cells of domestic production. Such a sequencer can be in demand in many clinical diagnostic laboratories.

https://store.nanoporetech.com/devices
PromethION disposable sequencer cells contain 9,000 sensor wells on chips. And the chips themselves are 3000-channel pico-amp current amplifiers. The flow of information cells issued by them from 24 (for PromethION 24) or from 48 (for PromethION 48) is so large that in its “raw” form it cannot be transferred to a regular external computer. Therefore, sequencers of this type are necessarily equipped with their own supercomputer. But if the sequencer will work with only one cell, then it can be connected to a regular gaming laptop with a Thunderbolt 3.0 port.
Alteration of free (ejected) chips from ONT to work with contact cells is not an insoluble problem. But sequencers based on the use of such chips can only work with ONT software. And the efforts that will have to be spent on reworking these programs can negate the benefits of using ready-made chips. And to develop such chips from scratch is too expensive.
Megaarder
This year, beta testing of the AXBIO (USA) nanoporous sequencer should begin. It uses chips with a million channels of current amplification, produced in Japan by the Israeli company Tower Semiconductor Ltd ... Expendable reagents and sales of these sequencers will be handled by the Chinese branch of the company AXBIO.

In the United States and the European Union, more than 7,000 patents are devoted to nanoporus sequencing, which creates unpredictable financial risks for local companies trying to engage in similar technologies. Apparently, this is the main reason for the high internationalization of this development. And allows Russian companies and laboratories to participate in the testing and improvement of the AXBIO technology without any restrictions, and also to produce (or assemble) such sequencers on its territory.
It is advisable to filter terabytes of raw data received by a nanoporous sequencer with megapixel chips (megarider). And transfer to the computer only the longest and high-quality reads. This will not only improve the quality of sequencing, but also reduce computer requirements, which is also important in mass production and widespread use of genomic sequencers. In addition, the excessive performance of mega-readers can reduce the duration of sequencing of the human genome from 2 ... 3 days (on ONT) to several hours.
The amount of technology
The success of ONT was facilitated by the organization of parallel conveyor optimization of the five basic elements of nanopore sequencing technology - a membrane, a nanopore, a motor (helicase), sequencing conditions, and decoding algorithms of read signals.

https://nanoporetech.com/resource-centre/videos/sub1000
The guaranteed shelf life of cells for nanoporous sequencing depends mainly on the stability of the bilayer lipid membrane. Now it is 6 weeks, although usually such membranes are destroyed within a few days. To increase their shelf life, hydrophobic polymer additives, surface-active block copolymers, cholesterol, antioxidants, cross-linking of fatty acids, etc. are used, and such modifications can interfere with the formation of ion channels. Therefore, to achieve stability BLM is very difficult.
This problem can be fundamentally solved by the formation of membranes in the working cells of the sequencer and the introduction of nanopores into them immediately before use. Independent preparation of cells for work will provide the possibility of their regeneration and repeated use. As for the shelf life, for dry cells it is practically unlimited, and the reagents necessary for their activation and regeneration can be stored for months.
The bottleneck in the technology of nanoprovod sequencing is at the same time the bottleneck of the nanopore, which determines the quality of the read information. Therefore, one of the main tasks of ONT was the search for membrane protein, which allows to obtain results of acceptable quality.
The first such protein was CsgG, an outer membrane protein found in many gram-negative bacteria. Its nine subunits form an ion channel in the membrane with wide entrance and exit vestibules, and one bottleneck. But it was possible to obtain sufficiently high-quality nanopores (R9, R9.4.1, R9.5.1) only after searching and analyzing hundreds of modified CsgG variants. This summer, cells with the next generation pores (R10 or R10b), characterized by increased accuracy of reading homopolymer repeats, should appear on sale.

https://nanoporetech.com/about-us/news/london-calling-clive-brown-and-team-plenary
The obligatory component of ONT's nanoporos sequencing technology is helicase, a molecular motor that unwinds double-stranded DNA and inhibits the promotion of single-stranded DNA through a nanopore. The maximum speed of DNA unwinding by such a motor now reaches 450 base pairs per second, although the speed of the electronic components of the MinION makes it possible to increase it to 1000 nucleotides per second. Theoretically, the acceleration of the work of helicase can double the performance of the MinION, but in practice this may lead to a deterioration in the quality of sequencing. Therefore, faster and more efficient helicases are unlikely to improve the performance of nanoporous sequencers such as MinION and PromethION.
The quality of sequencing depends on a variety of conditions - the magnitude of the voltage applied to the membrane, the electrical conductivity and the ionic composition of the reaction mixture, the amount and nature of impurities plugging the pores, the ability to clean the blocked pores by changing the polarity of the potential, the structure of the adapters used, etc., etc. Therefore, nanoporous sequencing requires careful optimization of all its parameters and compositions of working solutions (Run Conditions) that can adversely affect the quality of digitization of nucleotide followers awns. For example, it has recently been shown that reducing the concentration of ATP cleaved by helicase gradually reduces the accuracy of sequencing results, and now Oxfords are trying to eliminate this problem by introducing ATP regeneration system into the reaction mixture.
The fifth element of nanopor sequencing technology is artificial intelligence, built on recurrent neural networks (RNN) and on the use of some special algorithms and programs that improve the recognition of homogeneous repetitions (Flip-flop, Medaka). In July, the next release of the Guppi and MinKNOW programs should appear, in which the Oxfords promise to add the possibility of identifying methylated bases (5mC and 6mA). This is important not only for epigenomic studies, but also to improve the recognition of common bases.
The large volumes of information read by nanopor sequencer and the complexity of its processing impose increased requirements on the hardware of such software and hardware complexes. Therefore, not every computer is suitable for connecting MinION, GridION and PromethION put the latest generation of graphics accelerators, and Oxford Nanopore Technologies became one of the first buyers of a large batch of Jetson AGX Xavier (NVIDIA) modules designed for devices with artificial intelligence.
More recently, nanoporous technology was considered nothing more than a useful addition to more accurate fluorescent and semiconductor sequencing technologies. But a gradual increase in the accuracy of DNA reading, combined with a large length of readable sequences, allowed her, if not to get ahead of all competitors, then at least determine the main vector of the development of genomic sequencing technologies.

A lot of time may be needed to master all the elements of nanopor sequencing technology, but this is not the biggest problem. The main problem is the lack of targeted financing of such complex developments in Russia. True, the recently approved Federal Scientific and Technical Program for the Development of Genetic Technologies for 2019–2027 (section “Program Implementation Directions”) says that in the short term (3 ... 6 years) a prototype of the device will be developed for high-performance genomic sequencing. But to find this single line on 30 pages of text is not so easy. It is easier to agree with the Chinese on the purchase of AXBIO sequencers.
It is even easier to find craftsmen who can assemble the simplest nanorider “on their knees”. And then walk with an outstretched hand in search of investors who will agree to finance the development of domestic technology of nanoporous sequencing.
Source: https://habr.com/ru/post/455156/
All Articles