Using WebAssembly, we accelerated web application 20 times
This article discusses a case for accelerating a browser application by replacing JavaScript calculations with WebAssembly.
WebAssembly - what is it?
In short, this is a binary instruction format for a stack virtual machine. Often, Wasm (abbreviated name) is called a programming language, but it is not. The instruction format is executed in the browser along with JavaScript.
It is important that WebAssembly can be obtained by compiling source codes in languages such as C / C ++, Rust, Go. Here statistical typing and the so-called flat memory model are applied. The code, as stated above, is stored in a compact binary format, so it runs almost as fast as if the application was started using the command line. These capabilities have led to the rise in popularity of WebAssembly.
')
We remind: for all readers of "Habr" - a discount of 10,000 rubles when writing to any Skillbox course on the promotional code "Habr".
Skillbox recommends: Practical course "Mobile Developer PRO" .
At the moment, Wasm is used in many applications, from games like Doom 3 to web applications like Autocad and Figma. Wasm is also used in the realm of serverless computing.
This article gives an example of using Wasm to speed up an analytical web service. For clarity, we took a working application written in C, which will be compiled into WebAssembly. The result will be used to replace the underperforming JS plots.
Application transformation
The example will use the fastq.bio browser service, which is intended for geneticists. The tool allows to evaluate the quality of DNA sequencing (decoding).
Here is an example application in work:

The details of the process are not worth mentioning, since they are rather difficult for non-specialists, but if in brief, then the scientists on the above infographic can understand whether the DNA sequencing process went smoothly and what problems they had.
This service has alternatives, desktop programs. But fastq.bio allows you to speed up the work by visualizing the data. In most other cases, you need to be able to work with the command line, but not all geneticists have the necessary experience.
Everything works simply. At the entrance - the data presented in the form of a text file. This file is generated by specialized sequencing tools. The file contains a list of DNA sequences and a quality assessment for each nucleotide. The file format is .fastq, so the service got its name.
JavaScript implementation
The first user step when working with fastq.bio is to select the appropriate file. Using the File object, the application reads a random sample of data from a file and processes this packet. The task of JavaScript here is to perform simple string operations and count indicators. One of them is the number of nucleotides A, C, G and T on different DNA fragments.
After calculating the required indicators, they are visualized using Plotly.js, and the service begins to work with a new data sample. Fragmentation is done to improve the quality of the UX. If you work with all the data at once, the process will freeze for a while, since files with sequencing results take up hundreds of gigabytes of file space. The service also takes sections of data from 0.5 to 1 MB in size and works with them step by step, building graphic data.
Here's how it works:

In the red box there is an algorithm for string conversions for visualization. This is the most loaded part of the service in terms of computing. It is worth trying to replace it with Wasm.
We test WebAssembly
To assess the possibility of using the Wasm, the project team started looking for ready-made solutions for creating a QC-metric (QC - quality control) based on fastq files. The search was conducted among the tools written in C, C ++ or Rust, so that it was possible to port the code to WebAssembly. In addition, the tool should not be "raw", it required a service already proven by scientists.
As a result, the choice was made in favor of seqtk . The application is quite popular, it is open-source, the source language is C.
Before converting to Wasm, you should look at the seqtk compilation principle for the desktop. According to the Makefile, here’s what you need:
# Compile to binary $ gcc seqtk.c \ -o seqtk \ -O2 \ -lm \ -lz In principle, you can compile seqtk using Emscripten. If it is not there, we get along with the Docker image .
$ docker pull robertaboukhalil/emsdk:1.38.26 $ docker run -dt --name wasm-seqtk robertaboukhalil/emsdk:1.38.26 If you wish, you can collect it yourself , but it takes time.
Inside the container you can easily take emcc as an alternative to gcc:
# Compile to WebAssembly $ emcc seqtk.c \ -o seqtk.js \ -O2 \ -lm \ -s USE_ZLIB=1 \ -s FORCE_FILESYSTEM=1 Minimum changes:
Instead of outputting to the Emscripten binary file, .wasm and .js are used to generate the files, which is used to launch the WebAssemby module.
To support the zlib library, the USE_ZLIB flag is used. The library is distributed and ported to WebAssembly, and Emscripten includes it in the project.
The virtual file system Emscrippten is activated. This is a POSIX-like FS running in RAM inside the browser. When the page is updated, the memory is cleared.
To understand why a virtual file system is needed, it is worth comparing the method of running seqtk from the command line with the method of running the compiled module WebAssembly.
# On the command line $ ./seqtk fqchk data.fastq # In the browser console > Module.callMain(["fqchk", "data.fastq"]) Getting access to the virtual file system is necessary in order not to rewrite seqtk for string rather than file input. In this case, the data fragment is displayed as a data.fastq file in a virtual file system with a call to it main () seqtk.
Here is the new architecture:
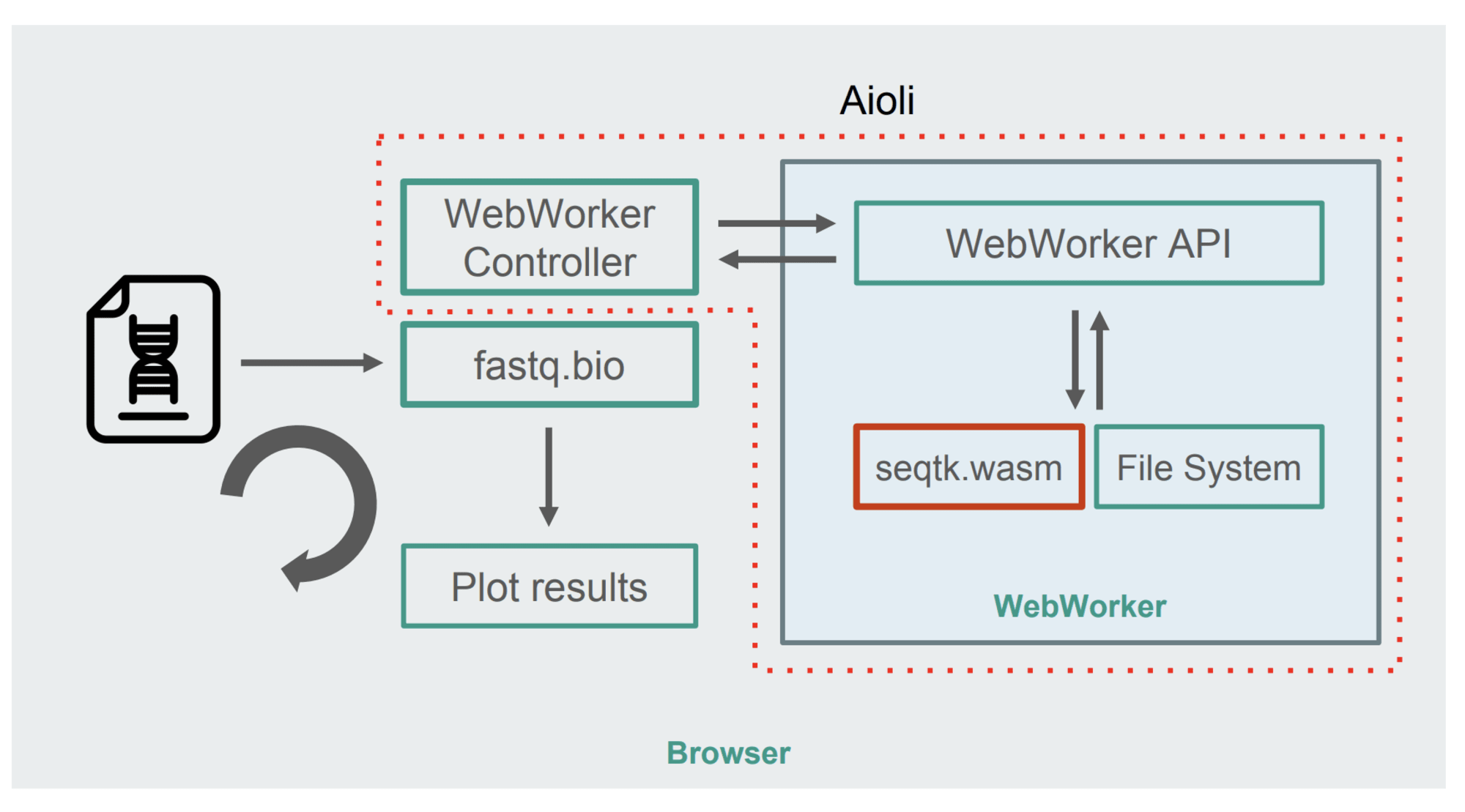
The figure demonstrates that instead of computing in the main browser thread, WebWorkers is used. This method allows you to perform calculations in the background thread, without compromising the responsiveness of the browser. Well, the WebWorker controller starts the Worker, controlling its interaction with the main thread.
The seqtk command is run by the Worker on the mounted file. After completion, the Worker returns the result as a Promise. When the message is received by the main thread, the result is used to update the graphs. And so in several iterations.
What about performance of WebAssembly?
In order to evaluate the performance change, the project team used the parameter for the number of read operations per second. The time for building interactive graphs is not taken into account, since JavaScript is used in both implementations.
When using the out-of-the-box solution, the performance increase was ninefold.

This is an excellent result, but as it turned out, it is possible to optimize it. The fact is that a large number of QC analysis results are not used by seqtk, so they can be deleted. If this is done, the result is 13 times better than JS.
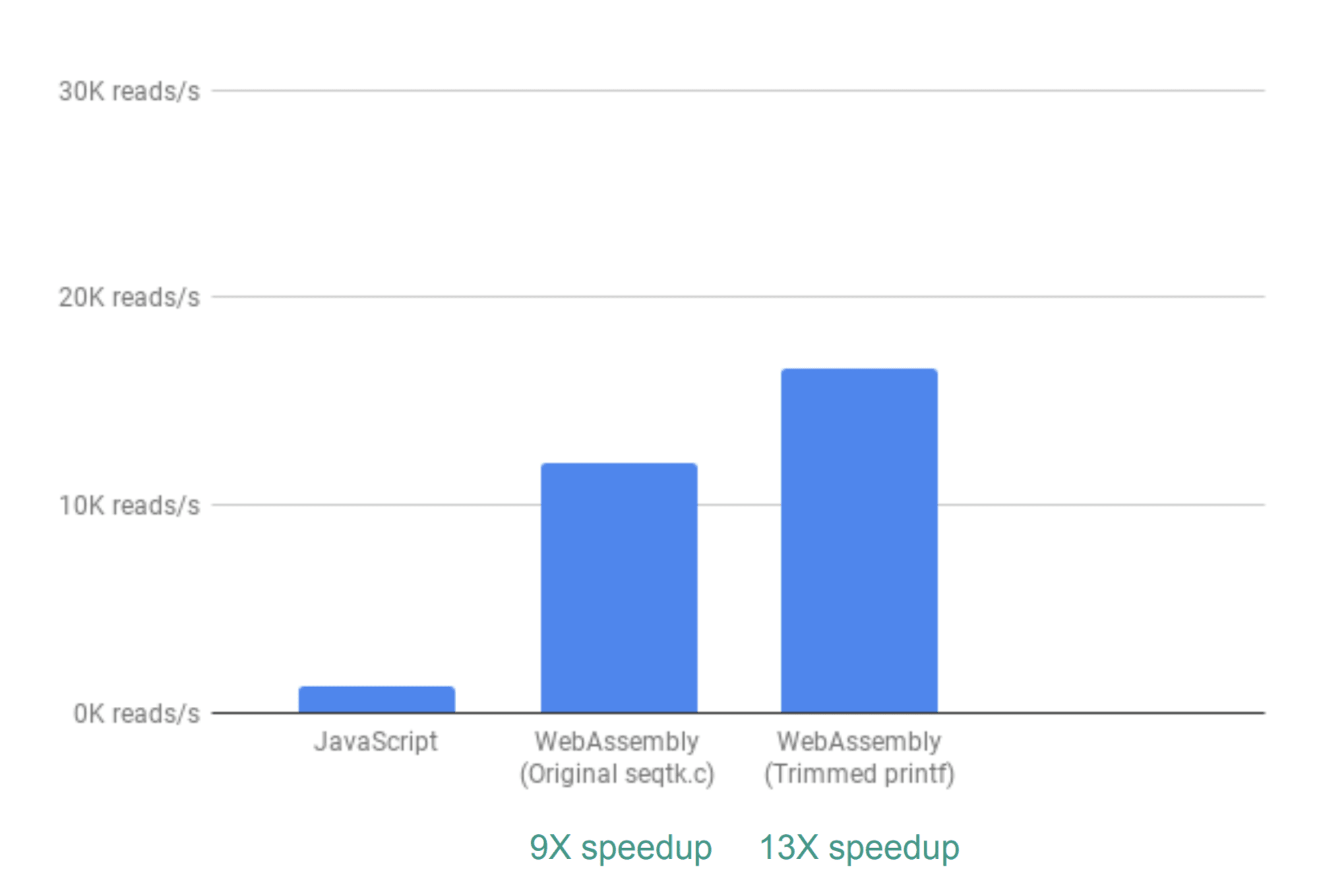
We managed to achieve it by simply commenting on the printf () commands.
But that's not all. The fact is that at this stage, fastq.bio receives the results of the analysis with calling various functions C. Each of them calculates its own set of characteristics, so each fragment of the file is read twice.
In order to get around this problem, it was decided to combine the two functions into one. As a result, productivity increased by 20 times.

It is worth noting that such an outstanding result can be achieved far from always. In some cases, performance drops, so it is worth evaluating each specific case.
As a conclusion, it can be said that Wasm really provides an opportunity to improve application performance, but you need to use it wisely.
Skillbox recommends:
- Two-year practical course "I am a web developer PRO" .
- Online course "C # developer with 0" .
- Practical annual course "PHP developer from 0 to PRO" .
Source: https://habr.com/ru/post/452190/
All Articles