"Hello"! The world's first automatic data storage in DNA molecules
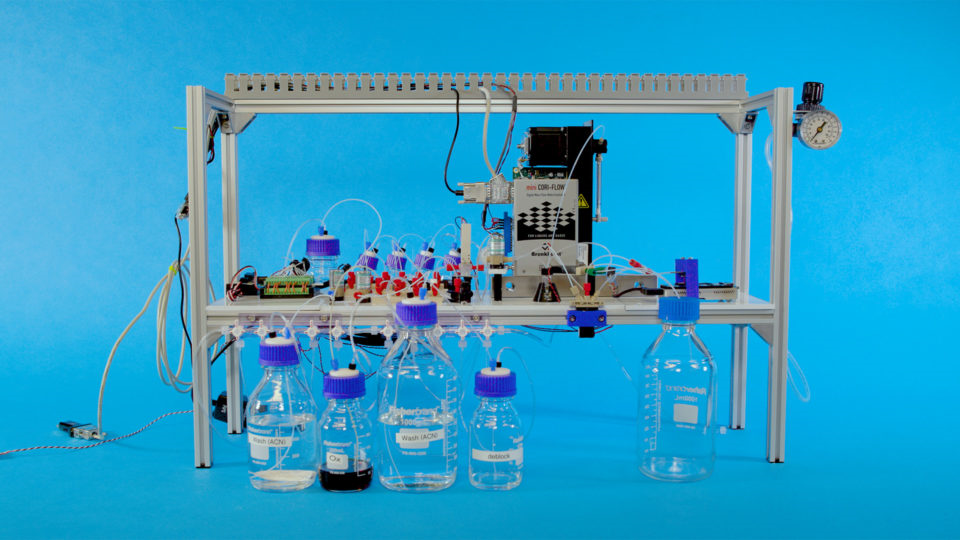
Researchers from Microsoft and the University of Washington demonstrated the first fully automated data storage system in artificially created DNA with the ability to read. This is a key step towards the transfer of new technology from research laboratories to commercial data centers.
The developers confirmed the concept with a simple test: successfully coded the word “hello” in fragments of a synthetic DNA molecule and transformed it back into digital data using a fully automated end-to-end system described in an article published March 21 in Nature Scientific Reports.
This article is on our website.
In DNA molecules, you can store digital information with a very high density, that is, in physical space, which is many orders of magnitude smaller than current data centers occupy. This is one of the most promising solutions for storing the huge amount of data that the world generates every day - from business records and videos with cute animals to medical images and images from space.
Microsoft is exploring ways to bridge the potential gap between the amount of data we produce and want to save, and our ability to store them. Such methods include the development of algorithms and molecular computing technologies for encoding data in artificial DNA . This would allow all the information stored in a large modern data center to fit into a space approximately equal to the size of several dice.
“Our main goal is to launch a system that will look to the end user almost like any other cloud storage system: the information is sent to the data center and stored there, and then just appear when the client needs it,” says Sr. Microsoft researcher Karin Strauss. “To do this, we had to prove that it makes practical sense from an automation point of view.”
Information is stored in synthetic DNA molecules created in the laboratory, and not in the DNA of humans or other living things, and can be encrypted before being sent to the system. Although complex machines, such as synthesizers and sequencers, already perform key parts of the process, many of the intermediate steps still required manual labor in a research lab. “This is not suitable for commercial use,” said Chris Takahashi, senior researcher at the Paul Allen School of Computer Science and Technology at the US University ( Paul G. Allen School of Computer Science & Engineering ).
“In the data center, people with pipettes cannot run; with this approach, the probability of human error is too high, it is too expensive and requires too much space,” explained Takahashi.
')
In order for this data storage method to make sense from a commercial point of view, it is necessary to reduce the costs of both DNA synthesis - the creation of fundamental building blocks with significant sequences, and the sequencing process, which is necessary for reading the stored information. Researchers say that there is a rapid development in this direction.
According to researchers from Microsoft, automation is another key part of this puzzle, allowing you to organize data storage on a commercial scale and make it more accessible.
Under certain conditions, DNA can exist much longer than modern archival storage tools that have been destroyed for decades. Some DNAs have been preserved in conditions that are far from ideal for tens of thousands of years — in the tusks of the mammoth and in the bones of early humans. This means that data can be stored in this way as long as humanity exists.
An automated DNA data storage system uses software developed by Microsoft and the University of Washington (UW). It converts the units and zeros of digital data into nucleotide sequences (A, T, C, and G), which are the “building blocks” of DNA. The system then uses inexpensive, mostly standard, laboratory equipment to supply the necessary liquids and reagents to the synthesizer, which collects the manufactured DNA fragments and places them in a storage tank.
When the system needs to extract information, it adds other chemicals to properly prepare DNA and uses microfluidic pumps to push fluids into parts of the system that read sequences of DNA molecules and convert them back to information that a computer can understand. The researchers say that the goal of the project was not to prove that the system could work quickly or cheaply, but simply to show that automation was possible.
One of the most obvious advantages of an automated DNA storage system is that it frees scientists to solve complex problems, allowing you not to waste time searching for bottles with reagents or monotonously adding liquid droplets to test tubes.
“Having an automated system for doing repetitive work allows laboratories to engage directly in research, develop new strategies to innovate more quickly,” said Microsoft researcher Bihlin Nguyen.
A team from the Molecular Information Systems Lab (MISL) has already demonstrated that it can store photos of seals, wonderful literary works, videos and archival DNA records and extract these files without error. To date, they were able to save 1 gigabyte of data in DNA, breaking the previous world record of 200 MB .
The researchers also developed methods to perform meaningful calculations , such as finding and extracting only images that have an apple or a green bicycle, using the molecules themselves, without converting the files back to digital format.
“It is safe to say that we are seeing the birth of a new type of computer system in which molecules are used to store data, and electronics are used for control and processing. This combination opens up very interesting opportunities for the future, ”said Louis Sété, a professor at Allen School of Washington University.
Unlike silicon-based computing systems, DNA-based storage and computing systems must use fluids to move molecules. But liquids are inherently different from electrons and require completely new technical solutions.
A team of Washington University, in collaboration with Microsoft, is also developing a programmable system that automates laboratory experiments, using the properties of electricity and water to move droplets on a grid of electrodes. A complete set of software and hardware, named Puddle and PurpleDrop , can mix, separate, heat or cool various liquids and perform laboratory protocols.
The goal is to automate laboratory experiments that are currently being done manually or with expensive liquid robots and reduce costs.
The following steps for the MISL team include the integration of a simple end-to-end automated system with technologies such as Purple Drop, as well as other technologies that allow you to search for DNA molecules. Researchers specially made their automated system modular, so that it could develop as new technologies appeared for synthesis, sequencing and working with DNA.
“One of the advantages of this system is that if we want to replace one of the parts with something new, more advanced or faster, we can simply plug in a new part,” said Nguyen. “It gives us more flexibility for the future.”
Top Image: Researchers at Microsoft and Washington University recorded and read the word " hello " using the first fully automated data storage system in DNA. This is a key step to transfer new technology from laboratories to commercial data centers.
Source: https://habr.com/ru/post/446496/
All Articles