R package tidyr and its new features pivot_long and pivot_wide
The tidyr package is included in the core of one of the most popular libraries in the R - tidyverse language .
The main purpose of the package is to bring data to a neat appearance.
On Habré there is already a publication dedicated to this package, but it dates back to 2015. And I want to tell you about the most urgent changes, which the author Headley Wickham told about it a few days ago.
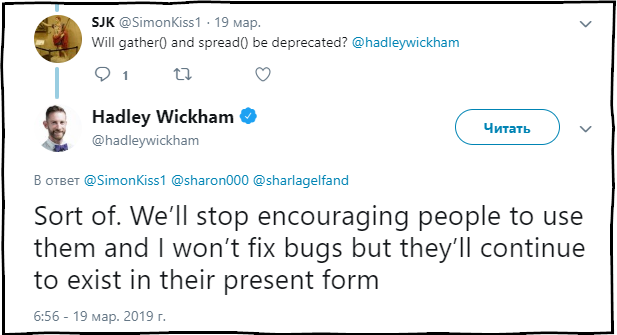
SJK : Will the functions gather () and spread () be obsolete?
Hadley Wickham : To some extent. We will stop recommending the use of these functions and correct errors in them, but they will continue to be present in the package in the current state.
Content
- TidyData concept
- Key features included in the tidyr package
- New concept of converting data from wide format to long and vice versa
- Installing the most current version of tidyr 0.8.3.9000
- Transition to new features
- A simple example of converting data from wide format to long
- Specs
- Specification using multiple values (.value)
- Convert date frames from long format to wide
- Some advanced examples of working with the new tidyr concept
- Conclusion
TidyData concept
The goal of tidyr is to help you bring the data to a so-called accurate look. Neat data is data, where:
- Each variable is in a column.
- Each observation is a string.
- Each value is a cell.
With data that is provided to tidy data it is much easier and more convenient to work when conducting an analysis.
Key features included in the tidyr package
tidyr contains a set of functions for transforming tables:
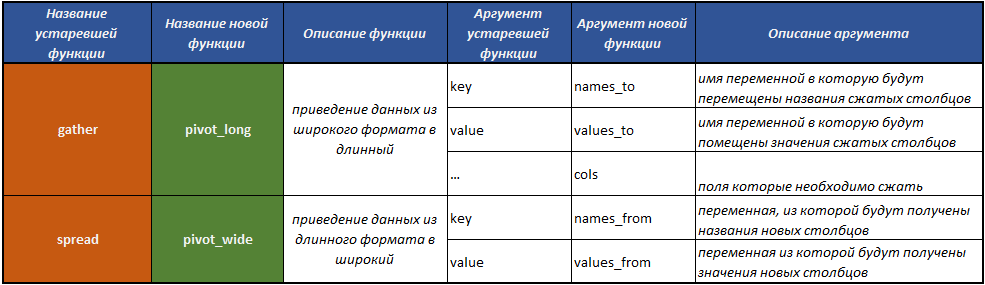
fill()- filling in the missing values in the column with previous values;separate()- splits one field into several through a separator;unite()- performs the operation of combining several fields into one, the inverse of the functionseparate();pivot_long()is a function that converts data from wide format to long;pivot_wide()is a function that converts data from a long format to a wide one. The operation is the inverse of the one performed by thepivot_long()function.gather()obsolete - a function that converts data from wide format to long;spread()obsolete - a function that converts data from a long format to a wide one. The operation is the reverse of the one that thegather()function performs.
New concept of converting data from wide format to long and vice versa
Previously, for this kind of transformation, the functions gather() and spread() . Over the years the existence of these functions, it became obvious that for most users, including the author of the package, the names of these functions and their arguments were not quite obvious, and caused difficulties in finding them and understanding which of these functions leads the frame from widely to long format, and vice versa.
In this connection, two new, important functions have been added to tidyr , which are designed to transform date frames.
The new functions pivot_long() and pivot_wide() were created under the impression of some of the functions from the cdata package created by John Mount and Nina Zumel.
Installing the most current version of tidyr 0.8.3.9000
To install the new, most current version of the package * tidyr 0.8.3.9000, in which new functions are available, use the following code.
devtools::install_github("tidyverse/tidyr")
At the time of this writing, these functions are only available in the dev version of the package on GitHub.
Transition to new features
In fact, it is easy to translate old scripts to work with new functions, for more understanding, I will take an example from the documentation of old functions and show how the same operations are performed using new pivot_*() functions.
Convert wide format to long.
# example library(dplyr) stocks <- data.frame( time = as.Date('2009-01-01') + 0:9, X = rnorm(10, 0, 1), Y = rnorm(10, 0, 2), Z = rnorm(10, 0, 4) ) # old stocks_gather <- stocks %>% gather(key = stock, value = price, -time) # new stocks_long <- stocks %>% pivot_long(cols = -time, names_to = "stock", values_to = "price") Convert long format to wide.
# old stocks_spread <- stocks_gather %>% spread(key = stock, value = price) # new stock_wide <- stocks_long %>% pivot_wide(names_from = "stock", values_from = "price") Since In the above examples of working with pivot_long() and pivot_wide() , there are no columns listed in the names_to and values_to arguments in their initial table stocks in quotes.
The table with which you will most easily deal with how to move to work with the new concept of tidyr .
Note from the author
All the text below is adaptive, I would even say the free translation of the vignette from the official website of the tidyverse library.
A simple example of converting data from wide format to long
pivot_long () - makes data sets longer by reducing the number of columns and increasing the number of rows.

To perform the examples presented in the article, you need to include the necessary packages:
library(tidyr) library(dplyr) library(readr) Suppose we have a table with survey results in which (among other things) people were asked about their religion and annual income:
#> # A tibble: 18 x 11 #> religion `<$10k` `$10-20k` `$20-30k` `$30-40k` `$40-50k` `$50-75k` #> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> #> 1 Agnostic 27 34 60 81 76 137 #> 2 Atheist 12 27 37 52 35 70 #> 3 Buddhist 27 21 30 34 33 58 #> 4 Catholic 418 617 732 670 638 1116 #> 5 Don't k… 15 14 15 11 10 35 #> 6 Evangel… 575 869 1064 982 881 1486 #> 7 Hindu 1 9 7 9 11 34 #> 8 Histori… 228 244 236 238 197 223 #> 9 Jehovah… 20 27 24 24 21 30 #> 10 Jewish 19 19 25 25 30 95 #> # … with 8 more rows, and 4 more variables: `$75-100k` <dbl>, #> # `$100-150k` <dbl>, `>150k` <dbl>, `Don't know/refused` <dbl> This table contains data about the religion of the respondents in rows, and the income level is scattered by column names. The number of respondents from each category is stored in cell values at the intersection of religion and income level. To bring the table to a neat, correct format, just use pivot_long() :
pew %>% pivot_long(cols = -religion, names_to = "income", values_to = "count") pew %>% pivot_long(cols = -religion, names_to = "income", values_to = "count") #> # A tibble: 180 x 3 #> religion income count #> <chr> <chr> <dbl> #> 1 Agnostic <$10k 27 #> 2 Agnostic $10-20k 34 #> 3 Agnostic $20-30k 60 #> 4 Agnostic $30-40k 81 #> 5 Agnostic $40-50k 76 #> 6 Agnostic $50-75k 137 #> 7 Agnostic $75-100k 122 #> 8 Agnostic $100-150k 109 #> 9 Agnostic >150k 84 #> 10 Agnostic Don't know/refused 96 #> # … with 170 more rows Arguments for the pivot_long() function
- The first argument, cols , describes which columns to merge. In this case, all columns except time .
- The names_to argument gives the name of the variable that will be created from the names of the columns we merged.
- values_to gives the name of the variable that will be created from the data stored in the cell values of the merged columns.
Specs
This is a new tidyr package functionality that was previously unavailable when working with obsolete functions.
The specification is a data frame, each row of which corresponds to one column in the new output date frame, and two special columns that begin with:
- .name contains the original column name.
- .value contains the name of the column in which the cell values will be entered.
The remaining specification columns reflect how the name of the compressible columns from .name will be displayed in the new column.
The specification describes the metadata stored in the column name, with one row for each column and one column for each variable combined with the column name. Probably now this definition seems confusing, but after considering a few examples, everything will become much clearer.
The point of the specification is that you can extract, modify and set new metadata to the converted data frame.
To work with specifications when converting a table from a wide format to a long one, use the pivot_long_spec() function.
As this function works, it takes any date frame, and forms its metadata in the manner described above.
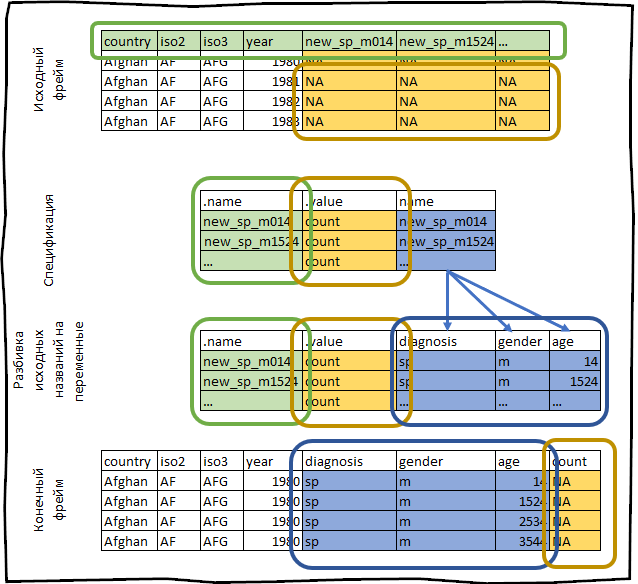
For example, let's take the who dataset, which is provided with the tidyr package. This data set contains information provided by the international health organization on the incidence of tuberculosis.
who #> # A tibble: 7,240 x 60 #> country iso2 iso3 year new_sp_m014 new_sp_m1524 new_sp_m2534 #> <chr> <chr> <chr> <int> <int> <int> <int> #> 1 Afghan… AF AFG 1980 NA NA NA #> 2 Afghan… AF AFG 1981 NA NA NA #> 3 Afghan… AF AFG 1982 NA NA NA #> 4 Afghan… AF AFG 1983 NA NA NA #> 5 Afghan… AF AFG 1984 NA NA NA #> 6 Afghan… AF AFG 1985 NA NA NA #> 7 Afghan… AF AFG 1986 NA NA NA #> 8 Afghan… AF AFG 1987 NA NA NA #> 9 Afghan… AF AFG 1988 NA NA NA #> 10 Afghan… AF AFG 1989 NA NA NA #> # … with 7,230 more rows, and 53 more variables We construct its specification.
spec <- who %>% pivot_long_spec(new_sp_m014:newrel_f65, values_to = "count") #> # A tibble: 56 x 3 #> .name .value name #> <chr> <chr> <chr> #> 1 new_sp_m014 count new_sp_m014 #> 2 new_sp_m1524 count new_sp_m1524 #> 3 new_sp_m2534 count new_sp_m2534 #> 4 new_sp_m3544 count new_sp_m3544 #> 5 new_sp_m4554 count new_sp_m4554 #> 6 new_sp_m5564 count new_sp_m5564 #> 7 new_sp_m65 count new_sp_m65 #> 8 new_sp_f014 count new_sp_f014 #> 9 new_sp_f1524 count new_sp_f1524 #> 10 new_sp_f2534 count new_sp_f2534 #> # … with 46 more rows The fields country , iso2 , iso3 are already variable. Our task is to flip the columns from new_sp_m014 to newrel_f65 .
The names of these columns store the following information:
- The prefix
new_says that the column contains data on new cases of tuberculosis, the current date frame contains information only on new diseases, therefore this prefix in the current context does not carry any meaning. sp/rel/sp/epdescribes how to diagnose a disease.m/fgender of the patient.014age range of the patient.
We can split these columns using the extract() function using a regular expression.
spec <- spec %>% extract(name, c("diagnosis", "gender", "age"), "new_?(.*)_(.)(.*)") #> # A tibble: 56 x 5 #> .name .value diagnosis gender age #> <chr> <chr> <chr> <chr> <chr> #> 1 new_sp_m014 count sp m 014 #> 2 new_sp_m1524 count sp m 1524 #> 3 new_sp_m2534 count sp m 2534 #> 4 new_sp_m3544 count sp m 3544 #> 5 new_sp_m4554 count sp m 4554 #> 6 new_sp_m5564 count sp m 5564 #> 7 new_sp_m65 count sp m 65 #> 8 new_sp_f014 count sp f 014 #> 9 new_sp_f1524 count sp f 1524 #> 10 new_sp_f2534 count sp f 2534 #> # … with 46 more rows Note that the .name column must remain unchanged, since this is our index in the column names of the original data set.
Gender and age ( gender and age columns) have fixed and known values, so it is recommended to convert these columns into factors:
spec <- spec %>% mutate( gender = factor(gender, levels = c("f", "m")), age = factor(age, levels = unique(age), ordered = TRUE) ) Finally, in order to apply the specification we created to the original date frame who we need to use the argument argument in the pivot_long() function.
who %>% pivot_long(spec = spec)
#> # A tibble: 405,440 x 8 #> country iso2 iso3 year diagnosis gender age count #> <chr> <chr> <chr> <int> <chr> <fct> <ord> <int> #> 1 Afghanistan AF AFG 1980 sp m 014 NA #> 2 Afghanistan AF AFG 1980 sp m 1524 NA #> 3 Afghanistan AF AFG 1980 sp m 2534 NA #> 4 Afghanistan AF AFG 1980 sp m 3544 NA #> 5 Afghanistan AF AFG 1980 sp m 4554 NA #> 6 Afghanistan AF AFG 1980 sp m 5564 NA #> 7 Afghanistan AF AFG 1980 sp m 65 NA #> 8 Afghanistan AF AFG 1980 sp f 014 NA #> 9 Afghanistan AF AFG 1980 sp f 1524 NA #> 10 Afghanistan AF AFG 1980 sp f 2534 NA #> # … with 405,430 more rows Everything that we have just done can be schematically depicted as follows:
Specification using multiple values (.value)
In the example above, the .value specification column contained only one value, in most cases this is the case.
But occasionally there may be a situation when you need to collect data in values from columns with different data types. Using the outdated function spread() would be quite difficult to do this.
The example below is borrowed from the vignette to the data.table package.
Let's create a training data frame.
family <- tibble::tribble( ~family, ~dob_child1, ~dob_child2, ~gender_child1, ~gender_child2, 1L, "1998-11-26", "2000-01-29", 1L, 2L, 2L, "1996-06-22", NA, 2L, NA, 3L, "2002-07-11", "2004-04-05", 2L, 2L, 4L, "2004-10-10", "2009-08-27", 1L, 1L, 5L, "2000-12-05", "2005-02-28", 2L, 1L, ) family <- family %>% mutate_at(vars(starts_with("dob")), parse_date) #> # A tibble: 5 x 5 #> family dob_child1 dob_child2 gender_child1 gender_child2 #> <int> <date> <date> <int> <int> #> 1 1 1998-11-26 2000-01-29 1 2 #> 2 2 1996-06-22 NA 2 NA #> 3 3 2002-07-11 2004-04-05 2 2 #> 4 4 2004-10-10 2009-08-27 1 1 #> 5 5 2000-12-05 2005-02-28 2 1 The created date frame in each row contains data about children of the same family. Families can have one or two children. For each child, data about the date of birth and the field are provided, and the data for each child is in separate columns, our task is to bring this data to the correct format for analysis.
Please note that we have two variables with information about each child: its gender and date of birth (columns with the dop prefix contain the date of birth, columns with the gender prefix contain the gender of the child). In the expected result, they should go in separate columns. We can do this by generating a specification in which the .value column has two different values.
spec <- family %>% pivot_long_spec(-family) %>% separate(col = name, into = c(".value", "child"))%>% mutate(child = parse_number(child)) #> # A tibble: 4 x 3 #> .name .value child #> <chr> <chr> <dbl> #> 1 dob_child1 dob 1 #> 2 dob_child2 dob 2 #> 3 gender_child1 gender 1 #> 4 gender_child2 gender 2 So, let's sort through the steps of the actions that are performed by the above code.
pivot_long_spec(-family)- create a specification that compresses all available columns, except the family column.separate(col = name, into = c(".value", "child"))- we divide the column .name , which contains the names of the original fields, by the underscore and enter the values in the columns .value and child .mutate(child = parse_number(child))- convert the values of the child field from text to numeric data type.
Now we can apply the received specification to the initial data frame and bring the table to the desired form.
family %>% pivot_long(spec = spec, na.rm = T) #> # A tibble: 9 x 4 #> family child dob gender #> <int> <dbl> <date> <int> #> 1 1 1 1998-11-26 1 #> 2 1 2 2000-01-29 2 #> 3 2 1 1996-06-22 2 #> 4 3 1 2002-07-11 2 #> 5 3 2 2004-04-05 2 #> 6 4 1 2004-10-10 1 #> 7 4 2 2009-08-27 1 #> 8 5 1 2000-12-05 2 #> 9 5 2 2005-02-28 1 We use the argument na.rm = TRUE , because the current form of the data forces us to create extra rows for nonexistent observations. Since family 2 has only one child, na.rm = TRUE ensures that family 2 will have one line in the output.
Convert date frames from long format to wide
pivot_wide() - is the inverse transformation, and vice versa increases the number of columns in the frame date by reducing the number of rows.
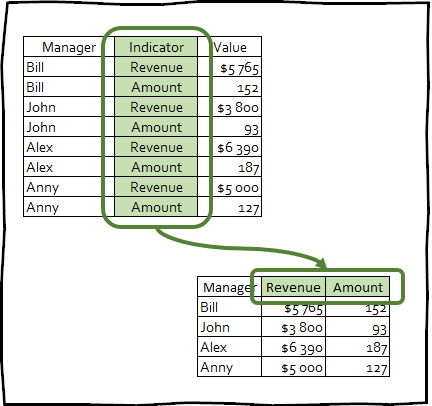
This kind of transformation is extremely rarely used to bring data to a neat appearance; nevertheless, this technique can be useful for creating summary tables used in presentations, or for integrating with any other tools.
In fact, the functions pivot_long() and pivot_wide() are symmetrical, and produce actions opposite to each other, that is: df %>% pivot_long(spec = spec) %>% pivot_wide(spec = spec) and df %>% pivot_wide(spec = spec) %>% pivot_long(spec = spec) returns the original df.
The simplest example of converting a table to a wide format
For the pivot_wide() function, we will use the fish_encounters dataset , which stores information on how various stations record the movement of fish along the river.
#> # A tibble: 114 x 3 #> fish station seen #> <fct> <fct> <int> #> 1 4842 Release 1 #> 2 4842 I80_1 1 #> 3 4842 Lisbon 1 #> 4 4842 Rstr 1 #> 5 4842 Base_TD 1 #> 6 4842 BCE 1 #> 7 4842 BCW 1 #> 8 4842 BCE2 1 #> 9 4842 BCW2 1 #> 10 4842 MAE 1 #> # … with 104 more rows In most cases, this table will be more informative and easy to use if you present the information for each station in a separate column.
fish_encounters %>% pivot_wide(names_from = station, values_from = seen)
fish_encounters %>% pivot_wide(names_from = station, values_from = seen) #> # A tibble: 19 x 12 #> fish Release I80_1 Lisbon Rstr Base_TD BCE BCW BCE2 BCW2 MAE #> <fct> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> #> 1 4842 1 1 1 1 1 1 1 1 1 1 #> 2 4843 1 1 1 1 1 1 1 1 1 1 #> 3 4844 1 1 1 1 1 1 1 1 1 1 #> 4 4845 1 1 1 1 1 NA NA NA NA NA #> 5 4847 1 1 1 NA NA NA NA NA NA NA #> 6 4848 1 1 1 1 NA NA NA NA NA NA #> 7 4849 1 1 NA NA NA NA NA NA NA NA #> 8 4850 1 1 NA 1 1 1 1 NA NA NA #> 9 4851 1 1 NA NA NA NA NA NA NA NA #> 10 4854 1 1 NA NA NA NA NA NA NA NA #> # … with 9 more rows, and 1 more variable: MAW <int> This data set records information only in cases when the fish was detected by the station, i.e. if any fish was not fixed by some station, then this data will not be in the table. This means that the output will be filled with NA.
However, in this case, we know that the absence of a record means that the fish was not noticed, so we can use the values_fill argument in the pivot_wide() function and fill these missing values with zeros:
fish_encounters %>% pivot_wide( names_from = station, values_from = seen, values_fill = list(seen = 0) ) #> # A tibble: 19 x 12 #> fish Release I80_1 Lisbon Rstr Base_TD BCE BCW BCE2 BCW2 MAE #> <fct> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> #> 1 4842 1 1 1 1 1 1 1 1 1 1 #> 2 4843 1 1 1 1 1 1 1 1 1 1 #> 3 4844 1 1 1 1 1 1 1 1 1 1 #> 4 4845 1 1 1 1 1 0 0 0 0 0 #> 5 4847 1 1 1 0 0 0 0 0 0 0 #> 6 4848 1 1 1 1 0 0 0 0 0 0 #> 7 4849 1 1 0 0 0 0 0 0 0 0 #> 8 4850 1 1 0 1 1 1 1 0 0 0 #> 9 4851 1 1 0 0 0 0 0 0 0 0 #> 10 4854 1 1 0 0 0 0 0 0 0 0 #> # … with 9 more rows, and 1 more variable: MAW <int> Generating a column name from multiple source variables
Imagine that we have a table containing a combination of product, country and year. To generate a test frame date, you can run the following code:
df <- expand_grid( product = c("A", "B"), country = c("AI", "EI"), year = 2000:2014 ) %>% filter((product == "A" & country == "AI") | product == "B") %>% mutate(value = rnorm(nrow(.))) #> # A tibble: 45 x 4 #> product country year value #> <chr> <chr> <int> <dbl> #> 1 A AI 2000 -2.05 #> 2 A AI 2001 -0.676 #> 3 A AI 2002 1.60 #> 4 A AI 2003 -0.353 #> 5 A AI 2004 -0.00530 #> 6 A AI 2005 0.442 #> 7 A AI 2006 -0.610 #> 8 A AI 2007 -2.77 #> 9 A AI 2008 0.899 #> 10 A AI 2009 -0.106 #> # … with 35 more rows Our task is to extend the date frame so that one column contains data for each combination of product and country. To do this, it is enough to transfer to the names_from argument a vector containing the names of the fields being combined.
df %>% pivot_wide(names_from = c(product, country), values_from = "value") #> # A tibble: 15 x 4 #> year A_AI B_AI B_EI #> <int> <dbl> <dbl> <dbl> #> 1 2000 -2.05 0.607 1.20 #> 2 2001 -0.676 1.65 -0.114 #> 3 2002 1.60 -0.0245 0.501 #> 4 2003 -0.353 1.30 -0.459 #> 5 2004 -0.00530 0.921 -0.0589 #> 6 2005 0.442 -1.55 0.594 #> 7 2006 -0.610 0.380 -1.28 #> 8 2007 -2.77 0.830 0.637 #> 9 2008 0.899 0.0175 -1.30 #> 10 2009 -0.106 -0.195 1.03 #> # … with 5 more rows You can also apply specifications to the pivot_wide() function. But when pivot_wide() to pivot_wide() specification performs the opposite transformation to pivot_long() : the columns specified in .name are created using values from .value and other columns.
For this dataset, you can generate a custom specification if you want each possible combination of country and product to have its own column, and not just those that are present in the data:
spec <- df %>% expand(product, country, .value = "value") %>% unite(".name", product, country, remove = FALSE) #> # A tibble: 4 x 4 #> .name product country .value #> <chr> <chr> <chr> <chr> #> 1 A_AI A AI value #> 2 A_EI A EI value #> 3 B_AI B AI value #> 4 B_EI B EI value df %>% pivot_wide(spec = spec) %>% head() #> # A tibble: 6 x 5 #> year A_AI A_EI B_AI B_EI #> <int> <dbl> <dbl> <dbl> <dbl> #> 1 2000 -2.05 NA 0.607 1.20 #> 2 2001 -0.676 NA 1.65 -0.114 #> 3 2002 1.60 NA -0.0245 0.501 #> 4 2003 -0.353 NA 1.30 -0.459 #> 5 2004 -0.00530 NA 0.921 -0.0589 #> 6 2005 0.442 NA -1.55 0.594 Some advanced examples of working with the new tidyr concept
Accurate data using the example of a census of income and rent census data in the United States
The us_rent_income dataset contains information on average income and rent for each state in the USA for 2017 (the dataset is available in the tidycensus package).
us_rent_income #> # A tibble: 104 x 5 #> GEOID NAME variable estimate moe #> <chr> <chr> <chr> <dbl> <dbl> #> 1 01 Alabama income 24476 136 #> 2 01 Alabama rent 747 3 #> 3 02 Alaska income 32940 508 #> 4 02 Alaska rent 1200 13 #> 5 04 Arizona income 27517 148 #> 6 04 Arizona rent 972 4 #> 7 05 Arkansas income 23789 165 #> 8 05 Arkansas rent 709 5 #> 9 06 California income 29454 109 #> 10 06 California rent 1358 3 #> # … with 94 more rows In the form in which data is stored in us_rent_income dataset, working with them is extremely inconvenient, so we would like to create a data set with columns: rent , rent_moe , come , income_moe . There are many ways to create this specification, but the main thing is that we need to generate each combination of variable and estimate / moe values, and then generate the column name.
spec <- us_rent_income %>% expand(variable, .value = c("estimate", "moe")) %>% mutate( .name = paste0(variable, ifelse(.value == "moe", "_moe", "")) ) #> # A tibble: 4 x 3 #> variable .value .name #> <chr> <chr> <chr> #> 1 income estimate income #> 2 income moe income_moe #> 3 rent estimate rent #> 4 rent moe rent_moe Providing this pivot_wide() specification gives us the result we are looking for:
us_rent_income %>% pivot_wide(spec = spec)
#> # A tibble: 52 x 6 #> GEOID NAME income income_moe rent rent_moe #> <chr> <chr> <dbl> <dbl> <dbl> <dbl> #> 1 01 Alabama 24476 136 747 3 #> 2 02 Alaska 32940 508 1200 13 #> 3 04 Arizona 27517 148 972 4 #> 4 05 Arkansas 23789 165 709 5 #> 5 06 California 29454 109 1358 3 #> 6 08 Colorado 32401 109 1125 5 #> 7 09 Connecticut 35326 195 1123 5 #> 8 10 Delaware 31560 247 1076 10 #> 9 11 District of Columbia 43198 681 1424 17 #> 10 12 Florida 25952 70 1077 3 #> # … with 42 more rows The World Bank
Sometimes casting a data set to the desired form requires several steps.
Dataset world_bank_pop contains the data of the World Bank on the population of each country in the period from 2000 to 2018.
#> # A tibble: 1,056 x 20 #> country indicator `2000` `2001` `2002` `2003` `2004` `2005` `2006` #> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> #> 1 ABW SP.URB.T… 4.24e4 4.30e4 4.37e4 4.42e4 4.47e+4 4.49e+4 4.49e+4 #> 2 ABW SP.URB.G… 1.18e0 1.41e0 1.43e0 1.31e0 9.51e-1 4.91e-1 -1.78e-2 #> 3 ABW SP.POP.T… 9.09e4 9.29e4 9.50e4 9.70e4 9.87e+4 1.00e+5 1.01e+5 #> 4 ABW SP.POP.G… 2.06e0 2.23e0 2.23e0 2.11e0 1.76e+0 1.30e+0 7.98e-1 #> 5 AFG SP.URB.T… 4.44e6 4.65e6 4.89e6 5.16e6 5.43e+6 5.69e+6 5.93e+6 #> 6 AFG SP.URB.G… 3.91e0 4.66e0 5.13e0 5.23e0 5.12e+0 4.77e+0 4.12e+0 #> 7 AFG SP.POP.T… 2.01e7 2.10e7 2.20e7 2.31e7 2.41e+7 2.51e+7 2.59e+7 #> 8 AFG SP.POP.G… 3.49e0 4.25e0 4.72e0 4.82e0 4.47e+0 3.87e+0 3.23e+0 #> 9 AGO SP.URB.T… 8.23e6 8.71e6 9.22e6 9.77e6 1.03e+7 1.09e+7 1.15e+7 #> 10 AGO SP.URB.G… 5.44e0 5.59e0 5.70e0 5.76e0 5.75e+0 5.69e+0 4.92e+0 #> # … with 1,046 more rows, and 11 more variables: `2007` <dbl>, #> # `2008` <dbl>, `2009` <dbl>, `2010` <dbl>, `2011` <dbl>, `2012` <dbl>, #> # `2013` <dbl>, `2014` <dbl>, `2015` <dbl>, `2016` <dbl>, `2017` <dbl> Our goal is to create a neat data set, where each variable is in a separate column. It is unclear exactly what steps are needed, but we will start with the most obvious problem: the year is divided into several columns.
In order to fix this, you must use the pivot_long() function.
pop2 <- world_bank_pop %>% pivot_long(`2000`:`2017`, names_to = "year") #> # A tibble: 19,008 x 4 #> country indicator year value #> <chr> <chr> <chr> <dbl> #> 1 ABW SP.URB.TOTL 2000 42444 #> 2 ABW SP.URB.TOTL 2001 43048 #> 3 ABW SP.URB.TOTL 2002 43670 #> 4 ABW SP.URB.TOTL 2003 44246 #> 5 ABW SP.URB.TOTL 2004 44669 #> 6 ABW SP.URB.TOTL 2005 44889 #> 7 ABW SP.URB.TOTL 2006 44881 #> 8 ABW SP.URB.TOTL 2007 44686 #> 9 ABW SP.URB.TOTL 2008 44375 #> 10 ABW SP.URB.TOTL 2009 44052 #> # … with 18,998 more rows The next step is to consider the variable indicator.pop2 %>% count(indicator)
#> # A tibble: 4 x 2 #> indicator n #> <chr> <int> #> 1 SP.POP.GROW 4752 #> 2 SP.POP.TOTL 4752 #> 3 SP.URB.GROW 4752 #> 4 SP.URB.TOTL 4752 Where SP.POP.GROW is population growth, SP.POP.TOTL is the total population, and SP.URB. * same, but only for urban areas. Let's divide these values into two variables: area - a location (total or urban) and a variable containing actual data (population or growth):
pop3 <- pop2 %>% separate(indicator, c(NA, "area", "variable")) #> # A tibble: 19,008 x 5 #> country area variable year value #> <chr> <chr> <chr> <chr> <dbl> #> 1 ABW URB TOTL 2000 42444 #> 2 ABW URB TOTL 2001 43048 #> 3 ABW URB TOTL 2002 43670 #> 4 ABW URB TOTL 2003 44246 #> 5 ABW URB TOTL 2004 44669 #> 6 ABW URB TOTL 2005 44889 #> 7 ABW URB TOTL 2006 44881 #> 8 ABW URB TOTL 2007 44686 #> 9 ABW URB TOTL 2008 44375 #> 10 ABW URB TOTL 2009 44052 #> # … with 18,998 more rows Now we just have to divide the variable variable into two columns:
pop3 %>% pivot_wide(names_from = variable, values_from = value) #> # A tibble: 9,504 x 5 #> country area year TOTL GROW #> <chr> <chr> <chr> <dbl> <dbl> #> 1 ABW URB 2000 42444 1.18 #> 2 ABW URB 2001 43048 1.41 #> 3 ABW URB 2002 43670 1.43 #> 4 ABW URB 2003 44246 1.31 #> 5 ABW URB 2004 44669 0.951 #> 6 ABW URB 2005 44889 0.491 #> 7 ABW URB 2006 44881 -0.0178 #> 8 ABW URB 2007 44686 -0.435 #> 9 ABW URB 2008 44375 -0.698 #> 10 ABW URB 2009 44052 -0.731 #> # … with 9,494 more rows List of contacts
Last example, imagine that you have a contact list that you copied and pasted from a website:
contacts <- tribble( ~field, ~value, "name", "Jiena McLellan", "company", "Toyota", "name", "John Smith", "company", "google", "email", "john@google.com", "name", "Huxley Ratcliffe" ) , , , . , , ("name"), , , field “name”:
contacts <- contacts %>% mutate( person_id = cumsum(field == "name") ) contacts #> # A tibble: 6 x 3 #> field value person_id #> <chr> <chr> <int> #> 1 name Jiena McLellan 1 #> 2 company Toyota 1 #> 3 name John Smith 2 #> 4 company google 2 #> 5 email john@google.com 2 #> 6 name Huxley Ratcliffe 3 , , :
contacts %>% pivot_wide(names_from = field, values_from = value) #> # A tibble: 3 x 4 #> person_id name company email #> <int> <chr> <chr> <chr> #> 1 1 Jiena McLellan Toyota <NA> #> 2 2 John Smith google john@google.com #> 3 3 Huxley Ratcliffe <NA> <NA> Conclusion
, tidyr , spread() gather() . pivot_long() pivot_wide() .
')
Source: https://habr.com/ru/post/444622/
All Articles