Iodide: Mozilla's interactive science editor.
Exploring the Lorenz attractor and then editing the code in Iodide
In the past ten years, there has been a real explosion of interest in “scientific computing” and “data science”, that is, the use of computational methods to search for answers to questions and to analyze data in the natural and social sciences. We see the heyday of specialized PL, tools and methods that help scientists explore and understand data and concepts, as well as report their findings.
But today very few scientific tools use the full communication potential of modern browsers. The results of the data mining is not very convenient to view in the browser. Therefore, today Mozilla introduces Iodide , an experimental tool that helps scientists compile beautiful interactive documents using web technologies, all within an iterative workflow that is familiar to many.
It is not just a programming environment for creating interactive documents in the browser. Iodide tries to help with the workflow by linking the editor and the preview. This is different from the IDE style, which produces presentation documents like pdf (they are then separated from the source code). And different from the style of notebooks with cells that mix code and presentation elements. In Iodide, you see both a document that looks the way you want and easy access to the underlying code and editing environment.
')
Iodide is still in the alpha version, but in the Internet industry they say: "If you are not confused by the first version of your product, you are late with the launch . " Therefore, we decided to make a very early launch in the hope of receiving feedback from the community. We have a demo that you can try right now , but there are still many flaws (please do not use this alpha for important work!). We hope that you close your eyes on the shoals and understand the value of the concept itself, and your feedback will help you to understand in which direction we should move on.
How we came to iodide
Data Science in Mozilla
The science of data in Mozilla is almost entirely based on communications. Although we sometimes deploy data mining models directly in front of users, such as the browser extension recommendation mechanism, most of the time our experts analyze data to identify patterns and share information with engineers, product managers and management.
The science of data involves writing a large amount of code, but unlike traditional software development, our goal is to answer questions rather than create software. This usually comes down to creating some kind of report — a document, a chart, or interactive data visualization. Like everyone else, we in Mozilla study our data with the help of fantastic modern tools such as Jupyter and R-Studio . But when it comes time to share the results, we usually can’t just hand over the Jupyter’s notebook or R script to the “customer,” so often you have to copy key numbers and summary statistics into a Google document.
As it turned out, it is quite difficult to move from studying the data in the code to a digestible explanation and back. Research shows that this is a common problem . When one scientist reads someone else's report and wants to see the corresponding code, many problems arise: sometimes it is easy to track the code, sometimes not. If a specialist wants to experiment by changing the code, it is obviously still complicated. Another scientist may have your code, but the configuration is different on the machine, and the setup takes time.

Useful Duty Cycle in Data Science
Why is there so little web in science?
Against this background, at the end of 2017, I started a project on interactive data visualization in Mozilla. Nowadays, you can create such visualizations with the help of excellent libraries in Python, R, and Julia, but for my project you had to switch to Javascript. This meant getting out of the usual environment of data science. Modern web development tools are incredibly powerful, but extremely complex . Against my will, I had to create a full-fledged chain of Javascript tools for assembling with hot reloading of modules, but still it was impossible to find a normal editor that generates clean, readable web documents in a live, iterative workflow.
I began to wonder why there is no such tool — why there is no Jupyter counterpart for interactive web documents — and I wondered why almost no one uses Javascript for scientific computing. It seems there are three important reasons for this:
- Javascript itself in scientific circles has a controversial reputation as a slow and inconvenient language.
- Not many scientific computing libraries work in a browser or support Javascript.
- I found a shortage of scientific programming tools with fast iteration support and direct access to browser presentation capabilities.
This is a very big problem. But working in a browser has some real advantages for this “communicative” data science, which we do in Mozilla. Of course, the biggest advantage is that the browser has the most advanced and well-supported set of data visualization technologies: from DOM to WebGL , Canvas and WebVR .
Reflecting on the difficulties in communication mentioned above, another potential advantage occurred to me: in the browser, the final document need not be separated from the tool that created it. I wanted to make a tool for the iterative scientific work with web documents (a web application with specific functionality) ... and many of our tools were essentially web applications. For these small web application documents, why not link the document with the editor?
Thus, readers without technical training can view a beautiful document, and the scientist instantly switches to source code mode. Moreover, since the calculations take place in the browser's JS engine, the scientist can immediately start experimenting with the code. And all this without connecting to remote computing resources or installing any software.
Appearance of iodide
I began to discuss with my colleagues potential pros and cons of scientific computing in the browser, and during the conversations we noted other interesting trends.
Inside Mozilla there were many interesting demonstrations on WebAssembly , a new platform for running code written in languages other than Javascript in the browser. WebAssembly allows you to run programs at an incredible speed, in some cases close to the native binaries. At WASM, resource-intensive processes work without problems, even entire 3D game engines . Basically, you can compile the world's best C and C ++ numerical computation libraries for WebAssembly, wrap them into ergonomic JS APIs, as the SciPy Python project does. In the end, such projects already exist .
WebAssembly allows you to run code in a browser with almost no overhead
We also noticed that the Javascript community is ready to introduce a new syntax if it helps people solve their problems more effectively. It may be worth trying to emulate some of the key syntax elements that make numerical programming more comprehensible and flexible in MATLAB, Julia and Python - this is matrix multiplication, multidimensional slicing, broadcast translation operations, and so on. And again we found that many agree with us .
All of these prerequisites lead to the question: how suitable is the web platform for scientific computing? At a minimum, it can help with communication in the processes that we face in Mozilla (and which many in industry and academic circles face). With the constantly improving Javascript core and the ability to add syntax extensions for numerical programming, it is possible that JS itself will become more attractive to scientists. It seemed, WebAssembly allowed to use serious scientific libraries. The third leg of the chair is a web environment for creating scientific documents. We focused our experiments on this last element, which led us to Iodide.
Anatomy of iodide
Iodide is a tool that provides scientists with a familiar workflow for creating great interactive documents using the power of a web platform. The work is built in the form of "reports" - in fact, this is a web page that you fill with your content. Plus some tools for iteratively examining data and modifying a report to create a final document. Once it is ready, you can send the link directly to it. If colleagues and employees want to view the code, then with one click of the mouse switch to the study mode. If they want to experiment with the code and use it as a basis for their work, then one more click becomes a fork.
Then we will talk about some experimental ideas on how to make this workflow more flexible.
Report Mode and Study Mode
Iodide seeks to link research, explanation and collaboration in one place. Central to this is the ability to navigate between a beautiful report and a useful environment for iterative research with scientific calculations.
When you create a new Iodide notebook, you are in Explore View mode. This is a set of panels, including an editor for writing code, a console for viewing output data, a workspace viewer for exploring variables created during a session, and a “Report Preview” pane.
Editing Markdown Code in Iodide Study Mode
By clicking on the Report button in the upper right corner, you can expand the contents of the preview pane to the whole window. Readers who are not interested in technical details can focus on this presentation of the document, without delving into the code. When the reader enters the link to the report, the code runs automatically. To view the code, click the Explore button in the upper right corner. From there you can make a copy of the notebook for your own research.
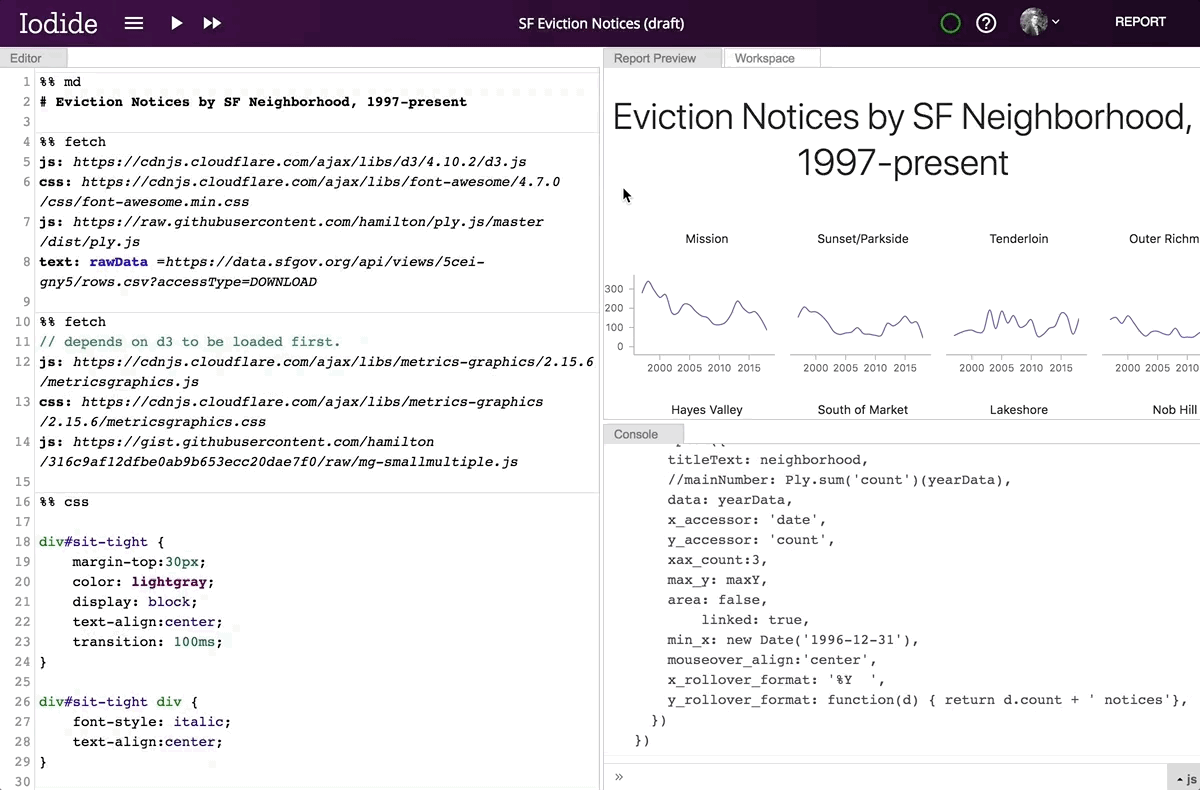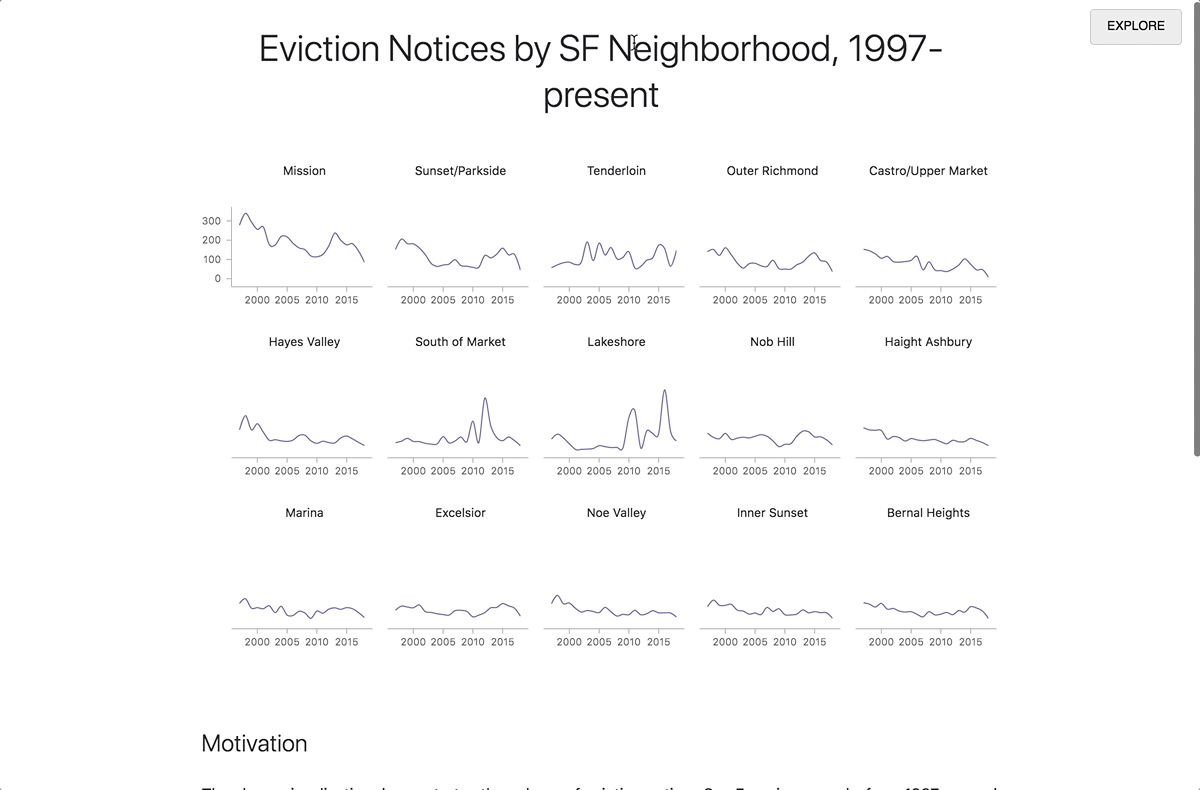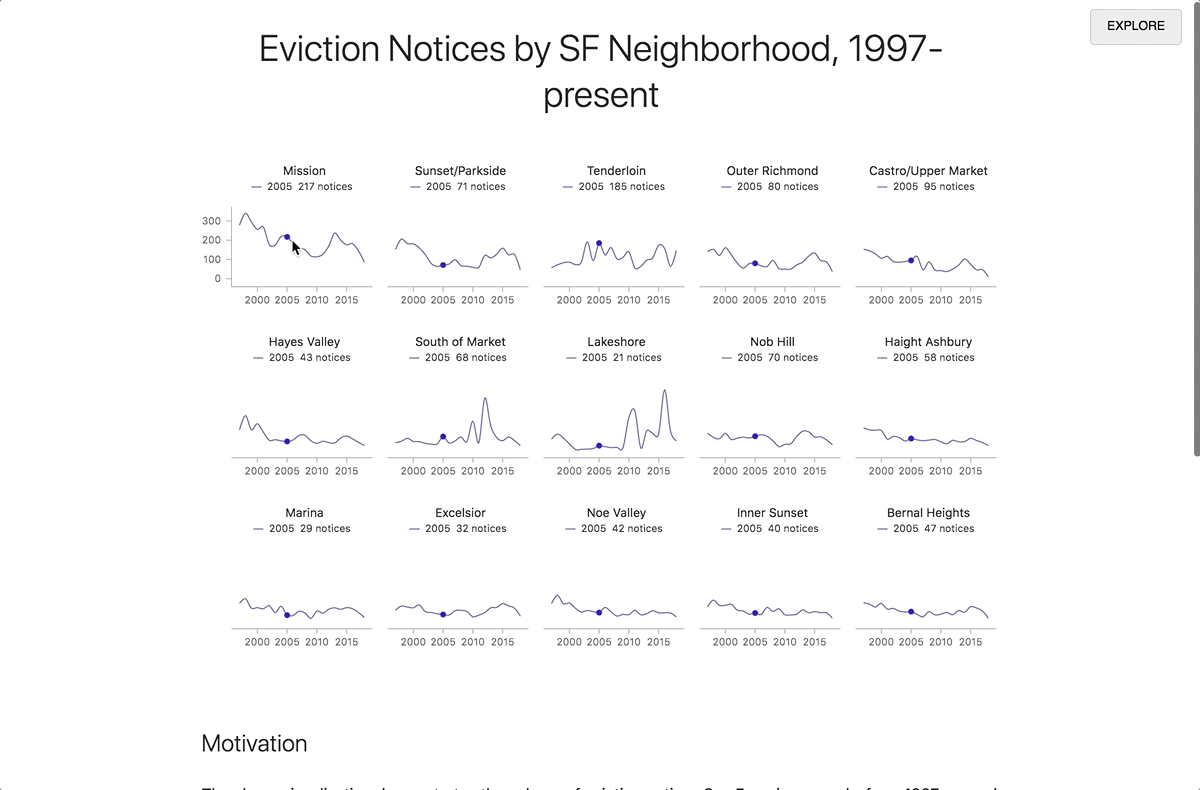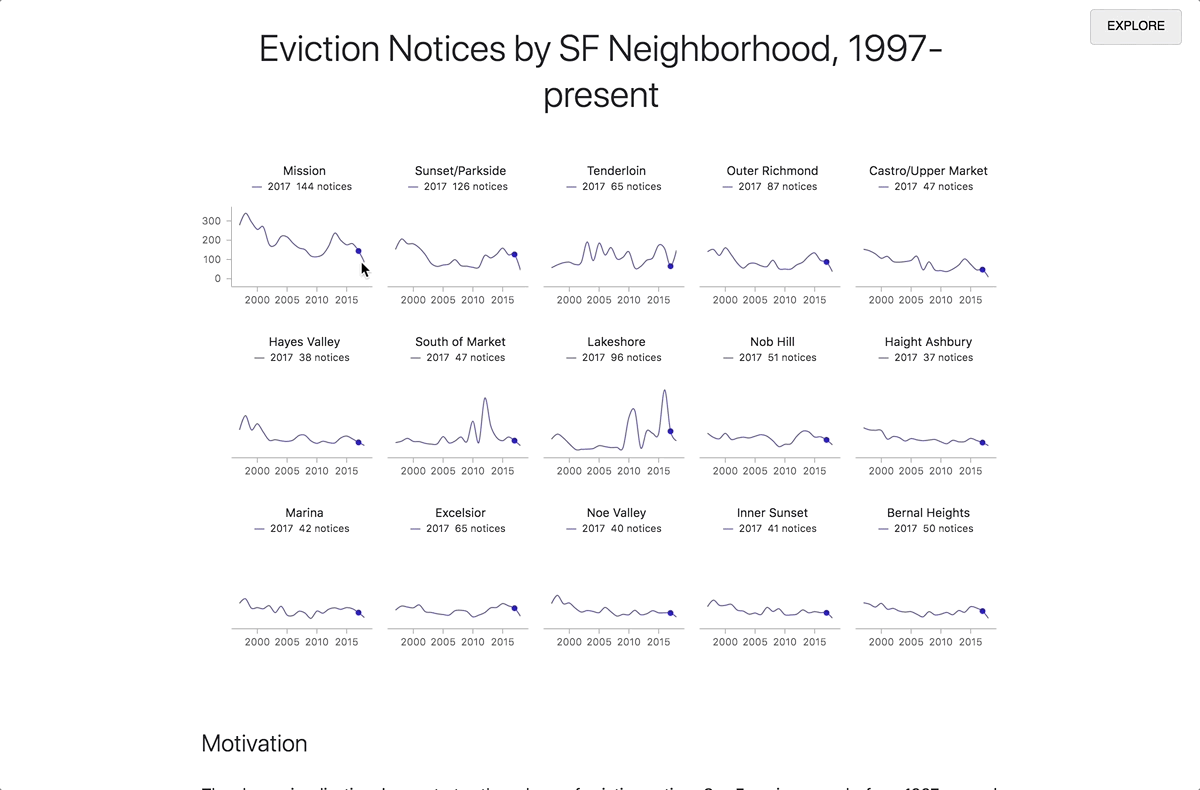
Transition from research mode to report mode
Whenever you share a link to an Iodide notebook, your colleague always gets access to both of these views. A clean, readable document is never separated from the underlying code and editing environment.
Live, interactive documents with the power of a web platform
Iodide documents live in a browser. This means that the computation engine is always available. Each document is a live interactive report with running code. Moreover, since the calculation takes place in the browser simultaneously with the presentation, there is no need to call the language backend in another process. Thus, interactive documents are updated in real time, opening the possibility of smooth 3D-visualizations . Low latency and high frame rates even meet VR requirements .
Contributor Devin Bailey examines his brain MRI data
Sharing and reproducibility
Reliance on the web simplifies a number of elements of the workflow, as compared to other tools. Sharing is implemented natively: the document and code are available at the same URL and you do not need to, say, insert a link to the script in the Google Docs footnotes. The computational core is a browser, and libraries are loaded with an HTTP request, like any script — no additional languages, libraries, or tools need to be installed. And since browsers provide compatibility, Notepad looks the same on all computers and OS.
To ensure collaboration, we created a fairly simple server where notepads are stored. There is an iodide.io public instance to experiment with Iodide and publish your work. But you can create a private instance behind the firewall (we in Mozilla do this for some internal documents). But it is important to note that the notebooks themselves are not tied to one Iodide server. If the need arises, it is easy to transfer work to another server or export the notepad as a package for sharing on other services, such as Netlify or GitHub Pages (for more details on exporting packages, see below in the section “What's next?”). Transferring computing to the client side allows us to focus on creating a truly excellent environment for exchange and collaboration, without having to allocate computing resources in the cloud.
Pyodide: Python science stack in browser
When we started thinking about how to make the web better for scientists, we focused on ways that can simplify working with Javascript, such as compiling existing research libraries in WebAssembly and packaging them into simple JS API. When we outlined the idea for WebAssembly developers at Mozilla , they suggested a more ambitious idea: if many scientists prefer Python, then step into their field — compile the Python science stack to run in WebAssembly.
We thought it sounded scary, that it would be a huge project and that it would never provide satisfactory performance ... but after two weeks Mike Droettbum had a working Python implementation running inside an Iodide notebook. Over the next few months, we added Numpy, Pandas and Matplotlib, the most used modules in the scientific ecosystem of Python. Thanks to the help of Cyril Smelkov and Roman Yurchak from Nexedi , Scipy and scikit-learn are supported. Since then, we continue to slowly add other libraries .
Running a Python interpreter inside a virtual machine Javascript adds performance overhead, but they are surprisingly small. Compared to the native code, in our tests the code runs 1-12 times slower in Firefox and 1-16 times slower in Chrome. Experience shows that productivity is enough for comfortable interactive research.
Matplotlib in the browser supports interactive features that are not available in static environments.
Moving Python to the browser creates magic workflows. For example, you can import and process data in Python, and then access the resulting objects from Javascript (in most cases, the conversion occurs automatically) to display them using JS libraries, such as d3 . Even more magically, you access the browser APIs from Python code, for example, to manipulate DOM without using Javascript .
Of course, much more can be said about Pyodide and it deserves a separate article - we will look at it in more detail next month.
JSMD (JavaScript MarkDown)
Like Jupyter and the R-Markdown mode in R, the Iodide editor allows you to freely alternate code and notes, breaking the code into pieces that change and run as separate units. Our implementation of this idea corresponds to the implementation of R Markdown and the “cell mode” in MATLAB: instead of the explicitly cell-based interface, the contents of the Iodide notepad are simply a text document that uses a special syntax to distinguish certain types of cells. We call this text format JSMD.
Modeled on MATLAB, code snippets begin with
%% , followed by a line indicating the language. We currently support fragments containing Javascript, CSS, Markdown (and HTML), Python, a special fragment of 'fetch', which simplifies the loading of resources, and plug-ins that extend the functionality of Iodide by adding new types of cells.We found this format very convenient. It simplifies the use of text tools, such as diff viewer and your own favorite text editor. You can perform standard text operations (cut / copy / paste) without having to learn the cell management commands. For more information, see the JSMD documentation .
What's next?
It is worth repeating that this is only the alpha version: we continue to polish the interface and fix errors. But in addition to this, there are a number of ideas for the following experiments. If any of these ideas seem particularly useful to you, let us know! Better yet, help develop!
Advanced collaboration features
As already mentioned, we have created a very simple backend that allows you to simply save work, view the work done by others, quickly fork and expand other people's notebooks. But these are only the first steps in a joint workflow.
Now we are considering three more great features:
- Comment Dreams in the style of Google Docs.
- The ability to propose changes in someone else's notepad through the fork / merge mechanism, as in GitHub.
- Simultaneous editing of notebooks, as in Google Docs.
At the moment, we set priorities roughly in this order, but if you decide to arrange them differently or you have other suggestions, do not hesitate to report it!
More languages!
We discussed with the R and Julia communities the possibility of compiling these languages into WebAssembly in order to use them in Iodide and other browser projects. At first glance, this is doable, but the implementation will be a bit more complicated than for Python. As in Python, some interesting workflows open up: for example, you can apply statistical models in R or solve differential equations in Julia and then display the results using the browser API. If you are involved in these languages, please let us know - in particular, we would like to receive help from FORTRAN and LLVM experts.
Export Notebooks
Early versions of Iodide were stand-alone executable HTML files that included both the JSMD code used in the analysis and the JS code to run Iodide itself, but we moved away from this architecture. Later experiments have convinced us that the benefits of working with the Iodide server outweigh the advantages of managing files on the local system. However, these experiments showed that you can make a standalone Iodide notepad executable by adding Iodide code along with any data and libraries you use into one large HTML file. It may be great, but it will be useful as a perfectly reproducible and archived snapshot.
Iodide browser extension for text editors
Although many scientists are accustomed to working in browser-based programming environments, some people will never give up their favorite text editor. We really want Iodide to go where it’s convenient for people to work, including those who prefer to enter code in another editor, but want to get access to the interactive and iterative functions provided by Iodide. To meet this need, we started thinking about creating a lightweight browser extension and some simple APIs to allow Iodide to communicate with the editors on the client side.
Feedback and help are welcome!
We are not trying to solve all the problems of scientific data and scientific calculations, and Iodide is not a universal solution. If you need to process terabytes of data on GPU clusters, Iodide is unlikely to help you. If you publish journal articles and you just need to write a document in LaTeX, then there are the best tools. If you don’t like the idea of transferring everything to the browser, there are no problems - there are many really amazing tools that you can use for science, and we thank you for it! We do not want to change someone's habits, and many scientists do not need web-based communications. Super! Do as you like!
But from the scientists who are doing or want to make content for the Internet, I would like to hear what tools you need in your work!
Please go to iodide.io , try this tool and leave a review (but again: keep in mind that the project is in the alpha version - please do not use it for any critical work, and remember that everything in alpha can change). You can fill out a short questionnaire , and tickets and bug reports on Github are also welcome. Feature requests and general thoughts should be left in our Google group or in Gitter .
If you want to participate in the creation of Iodide, the source code is published on Github . Iodide covers a wide range of software disciplines: from the development of modern interfaces and scientific computing to compilation and transfiguration, so there are many interesting things here!
Source: https://habr.com/ru/post/444596/
All Articles