Epigenetic Aging Biomarkers
The history of epigenetic biomarkers began in 2013. Then the pioneer in this direction, an expert in the field of genetics and biostatistics, an employee of the University of California at Los Angeles, Steve Horvath, presented his new revolutionary method for determining the biological age, called the "epigenetic clock." As the name implies, this method is based on changes in the epigenome, namely DNA methylation.
DNA methylation is one of the epigenetic mechanisms of gene expression regulation. In the course of methylation, the methyl group CH3-with special enzymes is attached to one of the bases of DNA, cytosine. As a result, 5-methylcytosine is formed and gene expression is inactivated - the transcription process is blocked. As is known today, DNA methylation is a dynamic process. It can change under the influence of external factors, is associated with the development of a number of pathologies and can be inherited by the next generations. Methylation plays a key role in the deactivation of foreign DNA, as well as in the processes of development and aging. Age-related changes in methylation, known as "epigenetic drift", are described. Thus, hypomethylation (demethylation) and the associated chromosomal instability are observed with age. In addition, the reverse process occurs during aging - hypermethylation of certain promoter regions, including certain tumor suppressor genes, which is associated with the development of pathologies [1]. In general, today it is believed that changing methylation plays a key role in aging.
Based on the fact that the chronological age is associated with predictable epigenome changes, hypo- and hypermethylation in many regions throughout the genome, the first generation of epigenetic biomarkers of aging based on DNA methylation was developed. In 2011, the first article by scientists at the University of California at Los Angeles, entitled “Epigenetic Age Predictor”, showed that DNA methylation has a clear correlation with age. In 2103, Steve Horvath identified 353 methylation sites using computer-aided learning techniques that were highly correlated with the chronological age of a person, which formed the basis of the first “epigenetic hours” or “Horvat hours” [2].
')
The importance of this discovery is difficult to overestimate. I must say that the success story of S. Horvat is full of not invented drama. According to the memoirs of Horvat himself, his article on the new epigenetic method of age measurement at first did not want to be accepted not in one of the journals. He constantly had to hear from the reviewers the same phrase, that "this is too good to be true." But Croat did not give up and devoted all his time to finalizing the new method: “I wrote in every free time, as if it was the last time to complete my article.” Just before the end of the work in the family of Horvat, a misfortune happened - his recently born daughter died. But this is not knocked out of the rut stubborn scientist. The last edition of his article was again not accepted in Genome Biology. And having received another critical comment from reviewers, Horvath, in his recollection, did three things that should not be done: “After reading the reviewers' comments, I spent the next 10 minutes doing three things that should never be done. First, I went to the fridge and drank three bottles of beer as fast as I could. Secondly, I went back to the computer and composed a letter to the editor. Third, I sent it. ” And this time fortune smiled at him, his article was accepted.

Steve Horvath.
How can an epigenetic clock be useful? According to the author of the discovery, this method can play a large role in evaluating the effectiveness of anti-aging interventions, as it allows to significantly shorten the waiting time for results. In addition, it is assumed that the sites of methylation identified by Horvath are not just markers, but also active participants in aging (at least some). What gives researchers a potential therapeutic target in the fight against aging and age-related pathologies. Also, this new method can be useful in criminal proceedings when it is necessary to establish an age by samples of fabric. But, of course, the most important "epigenetic clock" later acquired in predicting diseases and increasing the risk of mortality.
As it turned out, “epigenetic clocks” showed the value “zero” in embryonic stem and induced pluripotent cells, spermatozoa, egg cells and placental cells. And during the first 5 years of life - an accelerated course associated with the development of the body. By the year 21, the “epigenetic clock” gradually slowed down its course, and then went at a more or less uniform speed, changing its indicators under the influence of external factors. Moreover, it turned out that different tissues age at different speeds. The most susceptible to the aging process was the breast tissue in women. From the point of view of epigenetics, the brain is aging most slowly, and blood cells and bone tissue show slightly accelerated aging [3].
In the same 2013, an article by Chinese and American scientists was published, representing another version of the “epigenetic clock” - “Hannum clock”. Bioengineer of the University of California at San Diego Gregory Hannum and colleagues studied DNA methylation profiles of 450,000 CpG dinucleotides from human blood cells between the ages of 19 and 101, highlighting the 71 methylation sites most correlated with age [4]. The main difference between the “Hannum clock” and the “Horvath clock” is in their specificity: DNA samples are taken from blood cells, and not from any tissue, as in the Croat method.
The main value of the “epigenetic clock” became clear soon enough. Comparing their progress with chronological age has great predictive value for estimating the risk of all-cause mortality and the development of many pathologies. If the “epigenetic clock” is in a hurry, diseases arise, accelerated aging and shortening of the life span. If they are slower than chronological age, there are good chances for longevity. Studies have shown that the "epigenetic hours" have a high correlation with all-cause mortality and age-related diseases.
In 2015, Horvath and his colleagues conducted research to determine the relationship between the occurrence of lung cancer and indicators of epigenetic age. After analyzing the data of 2,029 people, it was found that the acceleration of epigenetic age was associated with an increased risk of lung cancer, and this relationship was stronger in smokers and those over 70 years of age: “The results showed that standardized indicators of acceleration of epigenetic age (IEAA) were significantly associated with the incidence of lung cancer (HR: 1.50, P = 3.4 × 10 -3). In addition, we have shown that the association can be even stronger among older people (70 years or older) or those who are currently smokers. Overall, our results indicate that IEAA may be a useful biomarker for assessing susceptibility to lung cancer from the point of view of biological aging ”[5].
In the same year, another group of researchers established a relationship between the “epigenetic age” and the risk of all-cause mortality in people over 60 years of age. For the analysis, data were taken from four studies (4,658 people in total), whose average age of participants was 79.1, 69.5, 66.3 and 72.9 years, respectively. The epigenetic age was determined by two methods: “Horvat clock” (based on 353 CpG methylation sites) and “Hannum clock” (based on 71 CpG methylation site). Both methods showed a strong correlation with each other, although the basic sets had matches only for 6 CpGs sites. The results of the study showed that the acceleration of the “epigenetic age” compared to the chronological one by 5 years increased the risk of mortality in humans by 16%. The authors concluded: “Accelerated aging indicators derived from DNA methylation are inherited factors that predict mortality regardless of health status, lifestyle factors, and known genetic factors. Therefore, it can be assumed that the predicted age of DNA methylation is an “epigenetic clock,” which measures the biological age, going together, but not always in parallel with the chronological age, and can give predictions of life expectancy ”[6].
In 2016, German oncologists conducted a study that described the relationship between accelerated epigenetic aging and mortality from cancer and cardiovascular diseases and all other causes. For their work, they used the “epigenetic clock” of Horvath and Hannum. The age of DNA methylation was assessed in a cohort of 1,863 elderly people in the ESTHER study, whose average age was 62.5 years. The results showed that the epigenetic age, exceeding the chronological age, was associated with a higher mortality. Acceleration by 5 years of “epigenetic age”, determined by the method of Horvat, gave an increase in mortality by 22%, and by 16% using the method of Hannum [7].
In 2016, a large international team of researchers led by Steve Horvath conducted a large-scale meta-analysis in which data from 13,089 people from three racial / ethnic groups were studied: whites, Latin Americans and African Americans. The epigenetic age here was also determined by two methods: “the clock of Horvath” and “the clock of Hannum”. This work showed that the acceleration of “epigenetic hours” for 1 year (compared to chronological age) increased the risk of all-cause mortality to 4%. Moreover, the opposite effect was observed: the slowing down of the “epigenetic hours” led to a decrease in the risk of mortality. In addition, the researchers recorded an interesting phenomenon: “We found that 5 percent of people have a faster biological age, which leads to a shorter lifespan. Accelerated aging increases the risk of death for these people by 50 percent in any adult age. ”[8, 9].
A number of external factors are also known that influence the course of the “epigenetic clock”. In 2017, Horvath and his colleagues described the relationship of diet, alcohol use, education and exercise with the course of epigenetic time. According to the authors themselves, their work confirmed a long-known truth: a diet with vegetables, fish and lean meat, moderate alcohol consumption, physical activity and education slow down the epigenetic time and contribute to the prolongation of life. Epigenome aging increased levels of insulin and glucose, C-reactive protein and triglycerides, as well as overweight and increased blood pressure. In the same study, the scientists did not find a positive effect of the anti-diabetic drug metformin on the course of epigenetic time [10]. In the same year, Finnish researchers once again demonstrated the relationship of obesity with accelerated epigenetic aging [11]
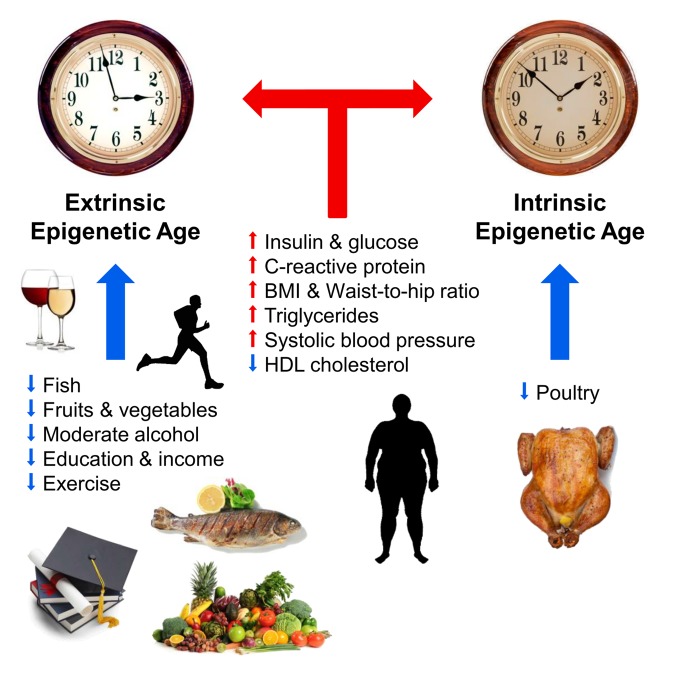
Fig. 1 Factors affecting the epigenetic age (from [10]).
Blue and red arrows show the factors that slow down and accelerate aging, respectively. The clock symbolizes the outer epigenetic clock (extrinsic epigenetic clock, the extended version of the Hannum method) and the inner epigenetic clock (intrinsic epigenetic clock, Horvath clock). Positively affect the course of epigenetic time (slow): fish, poultry, vegetables and fruits, high density lipoproteins, moderate alcohol, education and physical activity. Negatively affect the course of epigenetic time (accelerate): elevated levels of insulin, glucose, C-reactive protein, triglycerides, increased blood pressure, increased body weight and the wrong ratio of waist and hips.
Different teams of researchers conducted work that described the relationship of the epigenetic time with physical and cognitive functions, Down and Werner syndromes, HIV infection, Alzheimer's disease and menopause [12-17]. A clear relationship was also demonstrated between longevity and the slowing down of the “epigenetic hours” on the example of the Italian 100-year-old centenarians [18]. For a long time, Steve Croat did not succeed in predicting the accelerated course of biological age with the Hutchinson-Gilford progeria using his method. But this year this problem was solved: he and his colleagues created special “epigenetic clocks” based on fibroblasts, consisting of 391 CpG sites called “skin-blood clock” [19].
The next stage of S. Horvat’s work was the development of a more efficient biomarker of aging, with improved diagnostic capabilities that predict the risk of mortality from various causes and the development of age-related diseases. The disadvantage of the previous “epigenetic hours”, according to Horvat, was that the use of chronological age as a reference for determining age-related deviations may exclude CpG sites, whose methylation profiles do not exhibit strong, time-dependent changes. Instead, they show only a discrepancy between biological and chronological age. Thus, it is important not only to capture CpG, which reflect the difference with chronological time, but also those that demonstrate differences in risk and physiological status among persons of the same chronological age. In 2018, a new biomarker, called DNAm PhenoAge, was created.
At the first stage, the team of S. Horvat analyzed on a large sample of people, 9926 people from the NHANES III study, the relationship of 42 clinical biomarkers and chronological age with the risk of mortality. As a result, 9 biomarkers were identified to predict phenotypic age:
1. Albumin
2. Creatinine
3. Glucose
4. C-reactive protein
5. The percentage of lymphocytes
6. The average volume of red blood cells
7. The width of the distribution of red blood cells
8. The number of leukocytes
9. Alkaline phosphatase.
Then they validated selected biomarkers — they tested their model of phenotypic age on a different sample of people (6209 people). The audit showed an increase in phenotypic age with an increase in the risk of all-cause mortality: a one-year increase in the phenotypic age was associated with a 9% increase in the risk of all-cause mortality, a 9% increase in the risk of death from aging-related diseases, a 10% increase in the risk of death from heart disease. vascular diseases, a 7% increase in the risk of death from cancer, a 20% increase in the risk of death from diabetes and a 9% increase in the risk of death from respiratory diseases.
At the second stage of the study, the authors associated DNA methylation with a phenotypic age. Scientists have determined exactly which methylated GpC sites are associated with these 9 clinical biomarkers predicting a phenotypic age. They identified 513 CpG sites that predicted phenotypic age. A new epigenetic marker was named DNAm PhenoAge. The test showed a fairly high accuracy of the new biomarker: between 1998 and 2007, the mean change in DNAm PhenoAge was 8.51 years, while the mean change in clinical phenotypic age was 8.88 years.
After that, the researchers conducted a run-in of their new biomarker marker using data from 4 large studies, the Women's Health Initiative (n = 4207), the Framingham Heart Study (n = 2553), the Normative Aging Study (n = 657) and the Jackson Heart Study (n = 1747) The results showed that DNAm PhenoAge was significantly associated with the subsequent risk of mortality in all studies (regardless of chronological age): the one-year increase in DNAm PhenoAge is associated with an increased risk of all-cause mortality of 4.5%.
Genetic analysis of DNAm PhenoAge showed that the 513 CpG sites used in it have the same 41 CpGs with the Horvat clock and the same 6 CpGs with the Hannum clock. Five identical CpG were detected in all three epigenetic methods. 149 CpG from DNAm PhenoAge are located in areas of CpG dinucleotide clusters (CpG islands).
All the data obtained during the study by Steve Horvath and his team showed that the new biomarker has a great predictive value in determining the risks of the occurrence of age-related diseases and the risk of mortality. “ Using an innovative two-step process, we developed a new epigenetic biomarker of aging, DNAm PhenoAge, which greatly exceeded previous biomarkers in terms of predictions for various aging factors, including all-cause mortality, cancer, general health, physical functioning, and Alzheimer's disease. Although this biomarker was developed using data from whole blood, it strongly correlates with age in each tissue and cell under test. Based on in-depth transcriptional analysis in sorted cells, we found that an increased epigenetic age relative to chronological age is associated with an increase in the activation of pro-inflammatory and interferon pathways and a decrease in activation of transcriptional / translational mechanisms, response to DNA damage and mitochondrial signatures. In general, this single epigenetic biomarker of aging can encompass risks for a variety of different results in different tissues and cells and provide an insight into important pathways of aging [20].
Author: Alexey Rzheshevsky.
DNA methylation is one of the epigenetic mechanisms of gene expression regulation. In the course of methylation, the methyl group CH3-with special enzymes is attached to one of the bases of DNA, cytosine. As a result, 5-methylcytosine is formed and gene expression is inactivated - the transcription process is blocked. As is known today, DNA methylation is a dynamic process. It can change under the influence of external factors, is associated with the development of a number of pathologies and can be inherited by the next generations. Methylation plays a key role in the deactivation of foreign DNA, as well as in the processes of development and aging. Age-related changes in methylation, known as "epigenetic drift", are described. Thus, hypomethylation (demethylation) and the associated chromosomal instability are observed with age. In addition, the reverse process occurs during aging - hypermethylation of certain promoter regions, including certain tumor suppressor genes, which is associated with the development of pathologies [1]. In general, today it is believed that changing methylation plays a key role in aging.
Based on the fact that the chronological age is associated with predictable epigenome changes, hypo- and hypermethylation in many regions throughout the genome, the first generation of epigenetic biomarkers of aging based on DNA methylation was developed. In 2011, the first article by scientists at the University of California at Los Angeles, entitled “Epigenetic Age Predictor”, showed that DNA methylation has a clear correlation with age. In 2103, Steve Horvath identified 353 methylation sites using computer-aided learning techniques that were highly correlated with the chronological age of a person, which formed the basis of the first “epigenetic hours” or “Horvat hours” [2].
')
The importance of this discovery is difficult to overestimate. I must say that the success story of S. Horvat is full of not invented drama. According to the memoirs of Horvat himself, his article on the new epigenetic method of age measurement at first did not want to be accepted not in one of the journals. He constantly had to hear from the reviewers the same phrase, that "this is too good to be true." But Croat did not give up and devoted all his time to finalizing the new method: “I wrote in every free time, as if it was the last time to complete my article.” Just before the end of the work in the family of Horvat, a misfortune happened - his recently born daughter died. But this is not knocked out of the rut stubborn scientist. The last edition of his article was again not accepted in Genome Biology. And having received another critical comment from reviewers, Horvath, in his recollection, did three things that should not be done: “After reading the reviewers' comments, I spent the next 10 minutes doing three things that should never be done. First, I went to the fridge and drank three bottles of beer as fast as I could. Secondly, I went back to the computer and composed a letter to the editor. Third, I sent it. ” And this time fortune smiled at him, his article was accepted.

Steve Horvath.
How can an epigenetic clock be useful? According to the author of the discovery, this method can play a large role in evaluating the effectiveness of anti-aging interventions, as it allows to significantly shorten the waiting time for results. In addition, it is assumed that the sites of methylation identified by Horvath are not just markers, but also active participants in aging (at least some). What gives researchers a potential therapeutic target in the fight against aging and age-related pathologies. Also, this new method can be useful in criminal proceedings when it is necessary to establish an age by samples of fabric. But, of course, the most important "epigenetic clock" later acquired in predicting diseases and increasing the risk of mortality.
As it turned out, “epigenetic clocks” showed the value “zero” in embryonic stem and induced pluripotent cells, spermatozoa, egg cells and placental cells. And during the first 5 years of life - an accelerated course associated with the development of the body. By the year 21, the “epigenetic clock” gradually slowed down its course, and then went at a more or less uniform speed, changing its indicators under the influence of external factors. Moreover, it turned out that different tissues age at different speeds. The most susceptible to the aging process was the breast tissue in women. From the point of view of epigenetics, the brain is aging most slowly, and blood cells and bone tissue show slightly accelerated aging [3].
In the same 2013, an article by Chinese and American scientists was published, representing another version of the “epigenetic clock” - “Hannum clock”. Bioengineer of the University of California at San Diego Gregory Hannum and colleagues studied DNA methylation profiles of 450,000 CpG dinucleotides from human blood cells between the ages of 19 and 101, highlighting the 71 methylation sites most correlated with age [4]. The main difference between the “Hannum clock” and the “Horvath clock” is in their specificity: DNA samples are taken from blood cells, and not from any tissue, as in the Croat method.
The main value of the “epigenetic clock” became clear soon enough. Comparing their progress with chronological age has great predictive value for estimating the risk of all-cause mortality and the development of many pathologies. If the “epigenetic clock” is in a hurry, diseases arise, accelerated aging and shortening of the life span. If they are slower than chronological age, there are good chances for longevity. Studies have shown that the "epigenetic hours" have a high correlation with all-cause mortality and age-related diseases.
In 2015, Horvath and his colleagues conducted research to determine the relationship between the occurrence of lung cancer and indicators of epigenetic age. After analyzing the data of 2,029 people, it was found that the acceleration of epigenetic age was associated with an increased risk of lung cancer, and this relationship was stronger in smokers and those over 70 years of age: “The results showed that standardized indicators of acceleration of epigenetic age (IEAA) were significantly associated with the incidence of lung cancer (HR: 1.50, P = 3.4 × 10 -3). In addition, we have shown that the association can be even stronger among older people (70 years or older) or those who are currently smokers. Overall, our results indicate that IEAA may be a useful biomarker for assessing susceptibility to lung cancer from the point of view of biological aging ”[5].
In the same year, another group of researchers established a relationship between the “epigenetic age” and the risk of all-cause mortality in people over 60 years of age. For the analysis, data were taken from four studies (4,658 people in total), whose average age of participants was 79.1, 69.5, 66.3 and 72.9 years, respectively. The epigenetic age was determined by two methods: “Horvat clock” (based on 353 CpG methylation sites) and “Hannum clock” (based on 71 CpG methylation site). Both methods showed a strong correlation with each other, although the basic sets had matches only for 6 CpGs sites. The results of the study showed that the acceleration of the “epigenetic age” compared to the chronological one by 5 years increased the risk of mortality in humans by 16%. The authors concluded: “Accelerated aging indicators derived from DNA methylation are inherited factors that predict mortality regardless of health status, lifestyle factors, and known genetic factors. Therefore, it can be assumed that the predicted age of DNA methylation is an “epigenetic clock,” which measures the biological age, going together, but not always in parallel with the chronological age, and can give predictions of life expectancy ”[6].
In 2016, German oncologists conducted a study that described the relationship between accelerated epigenetic aging and mortality from cancer and cardiovascular diseases and all other causes. For their work, they used the “epigenetic clock” of Horvath and Hannum. The age of DNA methylation was assessed in a cohort of 1,863 elderly people in the ESTHER study, whose average age was 62.5 years. The results showed that the epigenetic age, exceeding the chronological age, was associated with a higher mortality. Acceleration by 5 years of “epigenetic age”, determined by the method of Horvat, gave an increase in mortality by 22%, and by 16% using the method of Hannum [7].
In 2016, a large international team of researchers led by Steve Horvath conducted a large-scale meta-analysis in which data from 13,089 people from three racial / ethnic groups were studied: whites, Latin Americans and African Americans. The epigenetic age here was also determined by two methods: “the clock of Horvath” and “the clock of Hannum”. This work showed that the acceleration of “epigenetic hours” for 1 year (compared to chronological age) increased the risk of all-cause mortality to 4%. Moreover, the opposite effect was observed: the slowing down of the “epigenetic hours” led to a decrease in the risk of mortality. In addition, the researchers recorded an interesting phenomenon: “We found that 5 percent of people have a faster biological age, which leads to a shorter lifespan. Accelerated aging increases the risk of death for these people by 50 percent in any adult age. ”[8, 9].
A number of external factors are also known that influence the course of the “epigenetic clock”. In 2017, Horvath and his colleagues described the relationship of diet, alcohol use, education and exercise with the course of epigenetic time. According to the authors themselves, their work confirmed a long-known truth: a diet with vegetables, fish and lean meat, moderate alcohol consumption, physical activity and education slow down the epigenetic time and contribute to the prolongation of life. Epigenome aging increased levels of insulin and glucose, C-reactive protein and triglycerides, as well as overweight and increased blood pressure. In the same study, the scientists did not find a positive effect of the anti-diabetic drug metformin on the course of epigenetic time [10]. In the same year, Finnish researchers once again demonstrated the relationship of obesity with accelerated epigenetic aging [11]

Fig. 1 Factors affecting the epigenetic age (from [10]).
Blue and red arrows show the factors that slow down and accelerate aging, respectively. The clock symbolizes the outer epigenetic clock (extrinsic epigenetic clock, the extended version of the Hannum method) and the inner epigenetic clock (intrinsic epigenetic clock, Horvath clock). Positively affect the course of epigenetic time (slow): fish, poultry, vegetables and fruits, high density lipoproteins, moderate alcohol, education and physical activity. Negatively affect the course of epigenetic time (accelerate): elevated levels of insulin, glucose, C-reactive protein, triglycerides, increased blood pressure, increased body weight and the wrong ratio of waist and hips.
Different teams of researchers conducted work that described the relationship of the epigenetic time with physical and cognitive functions, Down and Werner syndromes, HIV infection, Alzheimer's disease and menopause [12-17]. A clear relationship was also demonstrated between longevity and the slowing down of the “epigenetic hours” on the example of the Italian 100-year-old centenarians [18]. For a long time, Steve Croat did not succeed in predicting the accelerated course of biological age with the Hutchinson-Gilford progeria using his method. But this year this problem was solved: he and his colleagues created special “epigenetic clocks” based on fibroblasts, consisting of 391 CpG sites called “skin-blood clock” [19].
The next stage of S. Horvat’s work was the development of a more efficient biomarker of aging, with improved diagnostic capabilities that predict the risk of mortality from various causes and the development of age-related diseases. The disadvantage of the previous “epigenetic hours”, according to Horvat, was that the use of chronological age as a reference for determining age-related deviations may exclude CpG sites, whose methylation profiles do not exhibit strong, time-dependent changes. Instead, they show only a discrepancy between biological and chronological age. Thus, it is important not only to capture CpG, which reflect the difference with chronological time, but also those that demonstrate differences in risk and physiological status among persons of the same chronological age. In 2018, a new biomarker, called DNAm PhenoAge, was created.
At the first stage, the team of S. Horvat analyzed on a large sample of people, 9926 people from the NHANES III study, the relationship of 42 clinical biomarkers and chronological age with the risk of mortality. As a result, 9 biomarkers were identified to predict phenotypic age:
1. Albumin
2. Creatinine
3. Glucose
4. C-reactive protein
5. The percentage of lymphocytes
6. The average volume of red blood cells
7. The width of the distribution of red blood cells
8. The number of leukocytes
9. Alkaline phosphatase.
Then they validated selected biomarkers — they tested their model of phenotypic age on a different sample of people (6209 people). The audit showed an increase in phenotypic age with an increase in the risk of all-cause mortality: a one-year increase in the phenotypic age was associated with a 9% increase in the risk of all-cause mortality, a 9% increase in the risk of death from aging-related diseases, a 10% increase in the risk of death from heart disease. vascular diseases, a 7% increase in the risk of death from cancer, a 20% increase in the risk of death from diabetes and a 9% increase in the risk of death from respiratory diseases.
At the second stage of the study, the authors associated DNA methylation with a phenotypic age. Scientists have determined exactly which methylated GpC sites are associated with these 9 clinical biomarkers predicting a phenotypic age. They identified 513 CpG sites that predicted phenotypic age. A new epigenetic marker was named DNAm PhenoAge. The test showed a fairly high accuracy of the new biomarker: between 1998 and 2007, the mean change in DNAm PhenoAge was 8.51 years, while the mean change in clinical phenotypic age was 8.88 years.
After that, the researchers conducted a run-in of their new biomarker marker using data from 4 large studies, the Women's Health Initiative (n = 4207), the Framingham Heart Study (n = 2553), the Normative Aging Study (n = 657) and the Jackson Heart Study (n = 1747) The results showed that DNAm PhenoAge was significantly associated with the subsequent risk of mortality in all studies (regardless of chronological age): the one-year increase in DNAm PhenoAge is associated with an increased risk of all-cause mortality of 4.5%.
Genetic analysis of DNAm PhenoAge showed that the 513 CpG sites used in it have the same 41 CpGs with the Horvat clock and the same 6 CpGs with the Hannum clock. Five identical CpG were detected in all three epigenetic methods. 149 CpG from DNAm PhenoAge are located in areas of CpG dinucleotide clusters (CpG islands).
All the data obtained during the study by Steve Horvath and his team showed that the new biomarker has a great predictive value in determining the risks of the occurrence of age-related diseases and the risk of mortality. “ Using an innovative two-step process, we developed a new epigenetic biomarker of aging, DNAm PhenoAge, which greatly exceeded previous biomarkers in terms of predictions for various aging factors, including all-cause mortality, cancer, general health, physical functioning, and Alzheimer's disease. Although this biomarker was developed using data from whole blood, it strongly correlates with age in each tissue and cell under test. Based on in-depth transcriptional analysis in sorted cells, we found that an increased epigenetic age relative to chronological age is associated with an increase in the activation of pro-inflammatory and interferon pathways and a decrease in activation of transcriptional / translational mechanisms, response to DNA damage and mitochondrial signatures. In general, this single epigenetic biomarker of aging can encompass risks for a variety of different results in different tissues and cells and provide an insight into important pathways of aging [20].
Author: Alexey Rzheshevsky.
Bibliography
- Vayserman A.M., Voitenko V.P., Mekhova L.V. Epigenetic epidemiology of age-related diseases. Ontogenesis. 2011. 42, 1–21;
- Horvath S. DNA methylation of human tissues and cell types. Genome Biol. 2013. 14, R115.
- Josh Mitteldorf. Methylation Aging Clock: An Update. February 14, 2018.
- Hannum, G; Guinney, J; Zhao, L; Zhang, L; Hughes, G; Sadda, S; Klotzle, B; Bibikova, M; Fan, JB; Gao, Y; Deconde, R; Chen, M; Rajapakse, I; Friend, S; Ideker, T; Zhang, K (2013). Genome-wide methylation profiles reveal human aging rates. Mol cell. 49: 359-367.
- Morgan E. Levine, H. Dean Hosgood, Brian Chen, Devin Absher, Themistocles Assimes and Steve Horvath. DNA methylation age of blood choices. Aging (Albany NY). 2015 Sep; 7 (9): 690–700.
- Riccardo E Marioni, Sonia Shah, et al. DNA methylation age of blood prediction all-cause mortality in later life. Genome Biol. 2015; 16 (1): 25.
- Laura Perna, Yan Zhang, Ute Mons, Bernd Holleczek, Kai-Uwe Saum, and Hermann Brenner. Epigenetic acceleration of cancer, cardiovascular, and all-cause mortality. Clin Epigenetics. 2016; 8: 64.
- Brian H. Chen, Riccardo E. Marioni et al. DNA methylation-based measures of biological age: meta-analysis of prediction time to death (Albany NY). 2016 Sep; 8 (9): 1844-1859.
- Epigenetic clock predicts life expectancy. ScienceDaily. 28 September 2016.
- Quach A1, Levine ME1 et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY). 2017 Feb 14; 9 (2): 419-446.
- Nevalainen T, Kananen L, Marttila S, Jylhävä J, Mononen N, Kähönen M, Raitakari OT, Hervonen A, Jylhä M, Lehtimäki T, Hurme M. Clin Epigenetics. 2017 Feb 14; 9:20.
- Marioni RE, Shah S, McRae AF, Ritchie SJ, Muniz-Terrera G, Harris SE, Gibson J, Redmond P, Cox SR, Pattie A, Corley J, Taylor A, Murphy L, et al .. The epigenetic clock is correlated with the Lothian Birth Cohort 1936. Int J Epidemiol. 2015; 44: 1388–96.
- Horvath S, Garagnani P, Bacalini MG, Pirazzini C, Salvioli S, Gentilini D, Di Blasio AM, Giuliani C, Tung S, Vinters HV, Franceschi C. Accelerated epigenetic aging in Down syndrome. Aging Cell. 2015; 14: 491–95.
- Horvath S, Levine AJ. HIV-1 Infection Accelerates J Infect Dis. 2015; 212: 1563–73.
- Maierhofer A, Flunkert J, Oshima J, Martin GM, Haaf T, Horvath S. Accelerated epigenetic aging in Werner syndrome. Aging (Albany NY). 2017; 9: 1143–52.
- Levine ME, Lucky AT, Bennett DA, Hormones, and Alzheimer's disease related cognitive functioning. Aging (Albany NY). 2015; 7: 1198–211
- Levine ME, Lu AT, Chen BH, Hernandez DG, Singleton AB, Ferrucci L, Bandinelli S, Salfati E, Manson JE, Quach A, Kusters CD, Kuh D, Wong A, et al. Menopause accelerates biological aging. Proc Natl Acad Sci USA. 2016; 113: 9327–32.
- Horvath S, Pirazzini C, Bacalini MG, Gentilini D, Di Blasio AM, Delledonne M, Mari D, Arosio B, Monti D, Passarino G, De Rango F, D'Aquila P, Giuliani C, et al. Decreased epigenetic age of PBMCs from the Italian semi-supercentenarians and their offspring. Aging (Albany NY). 2015; 7: 1159–70.
- Steve Horvath, Junko Oshima et al. Hildeson Gilford Progeria Syndrome and ex vivo studies. Aging. Volume 10, Issue 7, pp 1758-75.
- 20. Morgan E. Levine, Ake T. Lu, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018 Apr; 10 (4): 573–591.
Source: https://habr.com/ru/post/428377/
All Articles