As the origin of man reflected in the genes
Recently, a video of The DNA Journey of the Danish company Momondo showed what effect the results of genetic testing on determining the composition of populations can have on people. On the air, people were told that they were far from such purebloods as they thought — and then emotions, tears and air tickets to their historic homeland. Such an analysis can be carried out not only in Europe and America. The Russian Atlas genetic test is able to determine the origin, population composition and percentage of Neanderthal genes. Behind all these studies is an interesting story that we will briefly tell today.
To understand the question, you need to understand how the inheritance of genetic information is arranged in principle.
')
Each person has 22 pairs of so-called autosomal chromosomes and one pair of sex chromosomes - XX or XY. In normal cell division, this set is doubled and then diverges into two cells, each of which receives an exact copy. Inside one organism, all these specimens are almost identical. Therefore, a genetic test can be performed on saliva, blood, hair follicles, cheek mucous membrane scraping, etc.
During the multiplication of organisms, the procedure becomes more complicated: each pair of chromosomes is doubled, and then working copies additionally exchange similar parts of the genome. Now, no matter what chromosome a new organism inherits, it will not be purely “grandmother’s” or “grandfather’s.” This process is called "crossing over": it is needed in order to make the genome more diverse. Then one complete set of chromosomes of the maternal organism meets with a set from the male organism, and a new gene is created.
At the same time, not all genetic material is subject to crossing-over - some elements are transferred to the daughter body unchanged. These are mitochondrial DNA (mtDNA), which is transmitted from the maternal organism to all children, and the Y chromosome, which is inherited through the male line, strictly from father to son.
Here lies the basis for paleogenetics: knowing the rate of mutation, scientists can calculate how far apart modern humans and their ancestors are. And the fun begins.

Now we see that two elements are inherited in isolation from each other: mtDNA is transmitted through the female line, and the Y chromosome is transmitted from father to son and nothing else. Knowing the sequence of these sections of the genome and the rate of mutation, scientists have calculated the dating of the common ancestors of mankind.
The woman was called Mitochondrial Eve. According to the latest data, he lived about 200-100 thousand years ago . Male - Y-chromosomal Adam - probably lived in the range from 450 to 50 thousand years ago . These two were not the only ancestors of humanity, just their brothers and sisters left no descendants in a straight (fatherly or maternal) line, but contributed differently to the history of humanity. They have enriched the nuclear genome — genetic information that is stored on autosomal and X chromosomes and makes up 99% of our genome.
At the same time, the Y-chromosomal Adam could be a descendant of other mitochondrial mothers, whose genealogical lines were interrupted.
Also, modern women are not all necessarily daughters of this Y-chromosomal Adam: their distant ancestors on the paternal line could be other Y-chromosomal fathers.
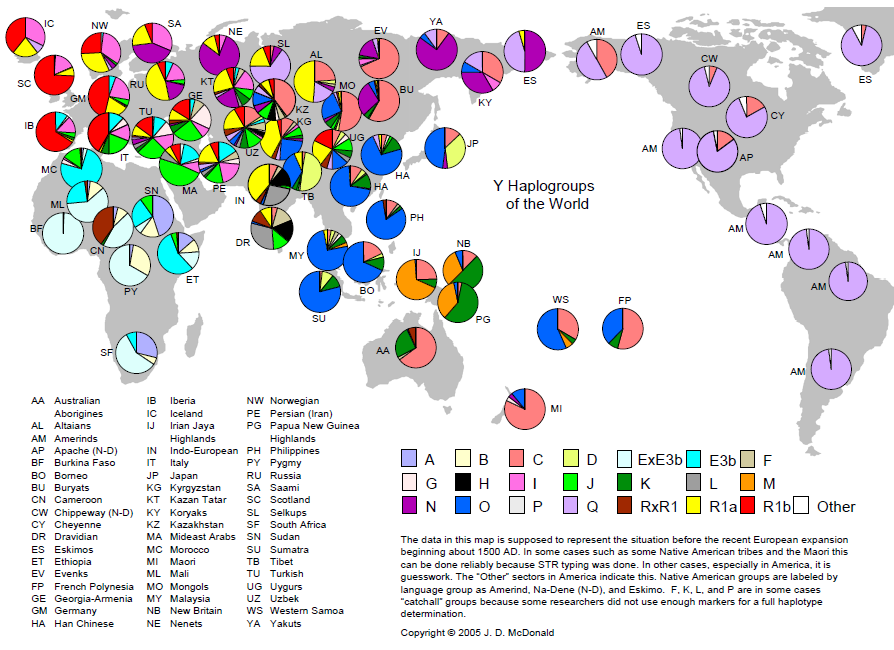
Studies of the genealogy of mtDNA and Y chromosomes led to the construction of a family tree, the various branches of which are called haplogroups. Members of one haplogroup are united by a common ancestor and one common chromosome. As the haplogroups opened (definitions of unique markers), scientists gave them letters of letters: A, B, C, and so on. However, after some time it turned out that the more ancient haplogroups were discovered later, and the harmonious numbering was broken.
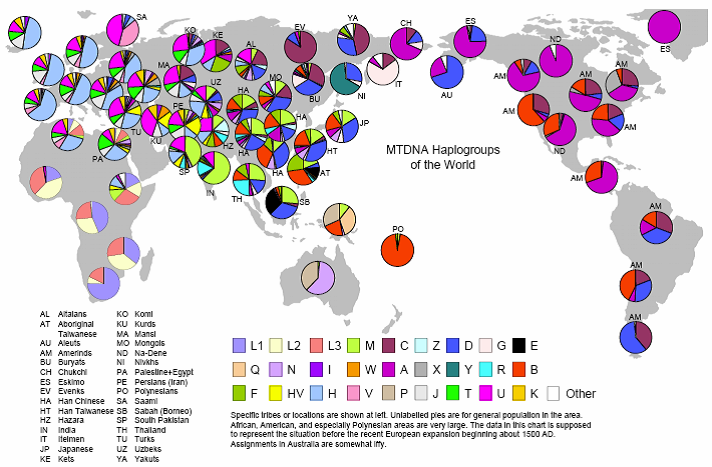
Haplogroups do not bear evolutionary advantages over each other: they are determined on the basis of neutral mutations, which, however, contribute to the likelihood of developing certain diseases.
The greatest diversity of haplogroups is observed in Africa, since the population of the rest of the globe is the descendants of those few surviving sapiens who left Africa and were able to survive in a new place ( bottle-neck effect ).
From the point of view of genetic testing, there are two methods for determining the haplogroup: by STR and SNP.
STR (short tandem repeat) - short tandem repeats that are in different loci (places) of the genome and are associated with the haplotype. Testing determines the number of repetitions of several nucleotides (for example, TAT) in significant areas of DNA for research. Based on the results a “map” of the number of repetitions is compiled The results are used not only to determine the haplogroup, but also to establish paternity.
SNP (single-nucleotide polymorphism) or snaps are single nucleotide mutations. The study finds point polymorphisms and, depending on which letter is written in a particular position, determines the haplogroup. The more snaps on a chip are assigned to the definition of a haplogroup, the higher the detail.
In the Atlas genetic test directly on the chip, 1000 positions are allocated for determining the maternal and paternal lines. We specifically included snaps to identify haplogroups characteristic of the Russian and European populations. African and Asian haplogroups will be defined with less detail).
Ethnic and national identity - concepts that are not applicable in biology. Therefore, genetics use the concept of "population" - "a collection of organisms of one species that have been living for a long time in one territory (occupying a certain range) and partially or completely isolated from individuals of other similar groups." As you understand, in the case of modern humanity there is no talk of complete isolation.

To determine the population composition, an algorithm is used that analyzes and clusters the genome samples of different inhabitants of the planet. Each cluster has a characteristic set of mutations (SNP-marker). Then scientists correlate clusters with data about research participants and distribute the names of populations. The resulting markers are used to determine the population composition in other genetic tests (and for the preparation of commercials).
To determine the population by marker, there are also different algorithms. In Atlas, we use ADMIXTURE . This algorithm is based on a component analysis of the user's genotype. We use machine learning: first we train the algorithm on the data of the latest studies of the peoples of the whole world, and then we calculate the population composition of the test participants. During training, the algorithm determines how many populations (component) can be distinguished based on the data that we contributed. After learning, the algorithm divides the sample of each user into components, which he calculated in the process of learning.
Now in the genetic test "Atlas" is determined by 11 populations. Among them are quite ordinary - Northern Europe, the Mediterranean, Southeast Asia, India, the Middle East, Africa, the Caucasus. And there are populations that are determined only by our test: Kamchatka, Eximosa, Yukagirs, Western Siberia.

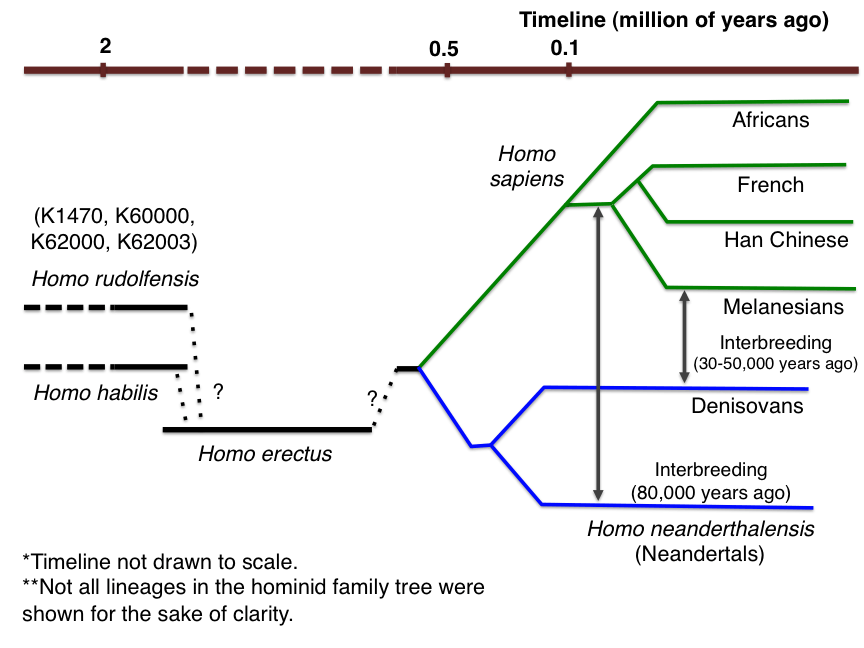
For a long time there was a theory that Neanderthals are the direct ancestors of modern man. But the latest discoveries and achievements of genetics speak in favor of another theory: we are not the direct descendants of Neanderthals, but rather their grand-nephews.
Neanderthals lived about 250–40 thousand years ago on the territory of modern Europe. They were well adapted to the cold climate: stocky, low-growth Neanderthals were folded in order to keep warm as much as possible. The volume of their brain is comparable to ours. Neanderthals hunted large animals, cared for the elderly, buried the dead, and even left two unique cultures ( Moustier and Micoc ) to humanity .
Everything was going well until 40 thousand years ago, Neanderthals began to die out, and their habitats were settled by Cro-Magnons (the direct ancestors of modern man). There are several theories that explain such changes: rapid and abrupt climate changes on the one hand, and competition with more successful Cro-Magnon scientists on the other.
For a long time, archaeological finds could not give an unequivocal answer to the question of what happened, and how different species - Cro-Magnon and Neanderthals - interacted with each other after the meeting. Paleogeneticists came to the aid of anthropologists.
In 1997, the Max Planck Institute for Evolutionary Anthropology Institute of Evolutionary Anthropology was opened. Under the supervision of the Swedish biologist Svente Paabo, the scientists read the mtDNA of a Neanderthal and concluded that it is very different from human. On this basis, Svente Paabo made the assumption that Neanderthals and people were divided about 500 thousand years ago and after that developed in isolation.

However, the work was continued. In 2006, the Neanderthal genome project ( Neanderthal genome project ) began its work. And already in 2010 a draft of the ancient genome was published. Now the Neanderthal genome is fully restored. The obtained data was compared with the genome of modern man (namely, with the “human” part of it, without taking into account those common places that coincide in humans and other animals).
The results of a comparative analysis showed that in the modern genome, from 1% to 4% of the Neanderthal genes are preserved (on average, about 2.5%). Neanderthal genes are equally found in all people except Africans - confusion occurred after Homo sapiens left Africa (once again).
If you are wondering what kind of haplogroup you have, which of the great people you are related to and how many Yukagirs were in your recent past - welcome to the Atlas . For attentive and patient readers 10% discount on the genetic test for promo code IAMSAPIENS (valid until August 1).
Effort on inheriting genetic information
To understand the question, you need to understand how the inheritance of genetic information is arranged in principle.
')
Each person has 22 pairs of so-called autosomal chromosomes and one pair of sex chromosomes - XX or XY. In normal cell division, this set is doubled and then diverges into two cells, each of which receives an exact copy. Inside one organism, all these specimens are almost identical. Therefore, a genetic test can be performed on saliva, blood, hair follicles, cheek mucous membrane scraping, etc.
During the multiplication of organisms, the procedure becomes more complicated: each pair of chromosomes is doubled, and then working copies additionally exchange similar parts of the genome. Now, no matter what chromosome a new organism inherits, it will not be purely “grandmother’s” or “grandfather’s.” This process is called "crossing over": it is needed in order to make the genome more diverse. Then one complete set of chromosomes of the maternal organism meets with a set from the male organism, and a new gene is created.
At the same time, not all genetic material is subject to crossing-over - some elements are transferred to the daughter body unchanged. These are mitochondrial DNA (mtDNA), which is transmitted from the maternal organism to all children, and the Y chromosome, which is inherited through the male line, strictly from father to son.
Here lies the basis for paleogenetics: knowing the rate of mutation, scientists can calculate how far apart modern humans and their ancestors are. And the fun begins.
Mitochondrial Eve and Y-chromosomal Adam

Now we see that two elements are inherited in isolation from each other: mtDNA is transmitted through the female line, and the Y chromosome is transmitted from father to son and nothing else. Knowing the sequence of these sections of the genome and the rate of mutation, scientists have calculated the dating of the common ancestors of mankind.
The woman was called Mitochondrial Eve. According to the latest data, he lived about 200-100 thousand years ago . Male - Y-chromosomal Adam - probably lived in the range from 450 to 50 thousand years ago . These two were not the only ancestors of humanity, just their brothers and sisters left no descendants in a straight (fatherly or maternal) line, but contributed differently to the history of humanity. They have enriched the nuclear genome — genetic information that is stored on autosomal and X chromosomes and makes up 99% of our genome.
At the same time, the Y-chromosomal Adam could be a descendant of other mitochondrial mothers, whose genealogical lines were interrupted.
Also, modern women are not all necessarily daughters of this Y-chromosomal Adam: their distant ancestors on the paternal line could be other Y-chromosomal fathers.
Haplogroups
Studies of the genealogy of mtDNA and Y chromosomes led to the construction of a family tree, the various branches of which are called haplogroups. Members of one haplogroup are united by a common ancestor and one common chromosome. As the haplogroups opened (definitions of unique markers), scientists gave them letters of letters: A, B, C, and so on. However, after some time it turned out that the more ancient haplogroups were discovered later, and the harmonious numbering was broken.

Haplogroups do not bear evolutionary advantages over each other: they are determined on the basis of neutral mutations, which, however, contribute to the likelihood of developing certain diseases.
The greatest diversity of haplogroups is observed in Africa, since the population of the rest of the globe is the descendants of those few surviving sapiens who left Africa and were able to survive in a new place ( bottle-neck effect ).

From the point of view of genetic testing, there are two methods for determining the haplogroup: by STR and SNP.
STR (short tandem repeat) - short tandem repeats that are in different loci (places) of the genome and are associated with the haplotype. Testing determines the number of repetitions of several nucleotides (for example, TAT) in significant areas of DNA for research. Based on the results a “map” of the number of repetitions is compiled The results are used not only to determine the haplogroup, but also to establish paternity.
SNP (single-nucleotide polymorphism) or snaps are single nucleotide mutations. The study finds point polymorphisms and, depending on which letter is written in a particular position, determines the haplogroup. The more snaps on a chip are assigned to the definition of a haplogroup, the higher the detail.
In the Atlas genetic test directly on the chip, 1000 positions are allocated for determining the maternal and paternal lines. We specifically included snaps to identify haplogroups characteristic of the Russian and European populations. African and Asian haplogroups will be defined with less detail).
Population composition
Ethnic and national identity - concepts that are not applicable in biology. Therefore, genetics use the concept of "population" - "a collection of organisms of one species that have been living for a long time in one territory (occupying a certain range) and partially or completely isolated from individuals of other similar groups." As you understand, in the case of modern humanity there is no talk of complete isolation.

To determine the population composition, an algorithm is used that analyzes and clusters the genome samples of different inhabitants of the planet. Each cluster has a characteristic set of mutations (SNP-marker). Then scientists correlate clusters with data about research participants and distribute the names of populations. The resulting markers are used to determine the population composition in other genetic tests (and for the preparation of commercials).
To determine the population by marker, there are also different algorithms. In Atlas, we use ADMIXTURE . This algorithm is based on a component analysis of the user's genotype. We use machine learning: first we train the algorithm on the data of the latest studies of the peoples of the whole world, and then we calculate the population composition of the test participants. During training, the algorithm determines how many populations (component) can be distinguished based on the data that we contributed. After learning, the algorithm divides the sample of each user into components, which he calculated in the process of learning.
Now in the genetic test "Atlas" is determined by 11 populations. Among them are quite ordinary - Northern Europe, the Mediterranean, Southeast Asia, India, the Middle East, Africa, the Caucasus. And there are populations that are determined only by our test: Kamchatka, Eximosa, Yukagirs, Western Siberia.

Neanderthals
For a long time there was a theory that Neanderthals are the direct ancestors of modern man. But the latest discoveries and achievements of genetics speak in favor of another theory: we are not the direct descendants of Neanderthals, but rather their grand-nephews.
Neanderthals lived about 250–40 thousand years ago on the territory of modern Europe. They were well adapted to the cold climate: stocky, low-growth Neanderthals were folded in order to keep warm as much as possible. The volume of their brain is comparable to ours. Neanderthals hunted large animals, cared for the elderly, buried the dead, and even left two unique cultures ( Moustier and Micoc ) to humanity .
Everything was going well until 40 thousand years ago, Neanderthals began to die out, and their habitats were settled by Cro-Magnons (the direct ancestors of modern man). There are several theories that explain such changes: rapid and abrupt climate changes on the one hand, and competition with more successful Cro-Magnon scientists on the other.
For a long time, archaeological finds could not give an unequivocal answer to the question of what happened, and how different species - Cro-Magnon and Neanderthals - interacted with each other after the meeting. Paleogeneticists came to the aid of anthropologists.
In 1997, the Max Planck Institute for Evolutionary Anthropology Institute of Evolutionary Anthropology was opened. Under the supervision of the Swedish biologist Svente Paabo, the scientists read the mtDNA of a Neanderthal and concluded that it is very different from human. On this basis, Svente Paabo made the assumption that Neanderthals and people were divided about 500 thousand years ago and after that developed in isolation.

However, the work was continued. In 2006, the Neanderthal genome project ( Neanderthal genome project ) began its work. And already in 2010 a draft of the ancient genome was published. Now the Neanderthal genome is fully restored. The obtained data was compared with the genome of modern man (namely, with the “human” part of it, without taking into account those common places that coincide in humans and other animals).
The results of a comparative analysis showed that in the modern genome, from 1% to 4% of the Neanderthal genes are preserved (on average, about 2.5%). Neanderthal genes are equally found in all people except Africans - confusion occurred after Homo sapiens left Africa (once again).

If you are wondering what kind of haplogroup you have, which of the great people you are related to and how many Yukagirs were in your recent past - welcome to the Atlas . For attentive and patient readers 10% discount on the genetic test for promo code IAMSAPIENS (valid until August 1).
Source: https://habr.com/ru/post/396089/
All Articles