Brief analysis of solutions in the field of SOC and the development of a neural network detector of anomalies in data networks

The article provides an analysis of solutions in the field of IDS and traffic processing systems, a brief analysis of attacks and analysis of the principles of IDS operation. After that, an attempt was made to develop a module for detecting anomalies in the network, based on the neural network method of analyzing network activity, with the following goals:
- Intrusion Detection
- Obtaining data on overloads and critical modes of network operation.
- Detection of network problems and network failures.
Goals and objectives of the article
The purpose of the article is to attempt to use the apparatus of neural networks in the applied area, mainly for the sake of studying the issue.
The task realizing the goal will be the construction of a neural network module for detecting deviations in the operation of a data transmission network from its normal modes.
Article structure
Research and justification:
- Types of intrusion detection systems.
- Brief market research for existing solutions.
- Brief study of works in the field of neural network solutions.
Development:
- Requirements for the module and its tasks.
- Design module.
- The implementation of the concept.
- Conclusion:
- Testing the concept.
- Opportunities for improvement.
- Used materials.
Types of Intrusion Detection Systems
There is a classical division of SOC in [11]:
- Network-level systems to which traffic from the router (Network-based) is diverted.
- Host-level systems that detect changes on a single machine, for example by analyzing logs or network activity (Host-based).
- Vulnerability-assessment based systems.
In this article I want to expand this division somewhat.
Actually, the purpose of any such system is to answer the question: are there any problems and which ones?
The decision is made on the basis of the data obtained.
Those. system tasks consist of:
- Receive data.
- Interpretation of the data.
- Presentation of the result.
Accordingly, all systems can be positioned by the values of the following features:
- Type of data collected.
- The method of obtaining data.
- Data interpretation method
- The method of presenting the result.
As a non-systemic characteristic, I consider the type of reaction to the result:
- Informational.
- Active.
In the first case, it informs interested parties.
In the second - active actions, such as blocking the range of addresses of the attacker.
On this basis, these systems are usually artificially divided into IDS and IPS.
This non-systemic characteristic, because I assume a partition of the system
on the "intelligence" and "power" parts. And any IDS can be included in the IPS.
Further, I will not return to this issue.
Classes of type of data collected
Following the classical division, I will introduce two classes and add a third one, so that it is possible to present the class as a "coordinate in configuration space" (just to quickly get all the permutations later):
- Data collected about the host.
- Data collected about the entire network.
- Hybrid system
Types of data collected about a host are data that relate to only one host and, in part, those who interact with it.
The analysis of such data allows to answer the question: "Is the attack going to this host?"
It is usually more convenient to collect this data directly at the site, but this is not necessary.
For example, network scanners can get a list of open ports on a particular host from the outside, without the ability to run code there.
This class includes the following types of data (each of which includes specific indicators collected):
- Network host activity.
- Network node settings.
- File information (lists and checksums, metadata, file actions, etc.).
- Process data
At the same time, nodes can be both workstations that do not intend to use them as servers providing services or servers.
I divide hosts into types, because I want to single out a separate case: the host can be specially made vulnerable in order to study attack methods (including the further training of neural networks) and identify attacking nodes.
It is possible to assume that any interaction with this node will be an attack attempt.
The data collected about the network is a complete picture of the network interaction.
As a rule, complete network data is not collected, because it is resource-intensive and it is believed that the intruder either cannot be inside the network, or he necessarily needs to communicate with the outside world (which, of course, is not necessarily the case, because it is possible to carry out an attack on a physically isolated network, for which there are techniques to "overcome the air gap" ).
In this case, the IDS analyzes the traffic going through the router, for which the router has a SPAN port from which the traffic is redirected to the IDS.
In principle, nothing prevents you from collecting data from the site where IDS is running, it is even useful and brings additional control.
It is also possible to collect network traffic on its nodes. But this makes the node's network adapter work in the mode of capturing all traffic, which is not usually assumed during normal operation, plus this is clearly redundant (the only option when it can be useful, with a big stretch, is distributed flow analysis).
Classes of data retrieval methods
Classes of data retrieval methods:
- Passive. The system does not directly affect network performance. It simply analyzes the traffic.
- Active. The system is trying to conduct a "reconnaissance in force", actively acting on the network, for example, with the aim of finding familiar signatures in the return traffic.
- Mixed. Both of the above classes are used.
Passive detection
With passive detection, the system simply monitors the situation. Most IDS use this class of methods. Host-level systems also typically use this class of methods. For example, they are not trying to remove the system file from the user and verify that it was deleted, but simply evaluate the compliance of the rights to this file with the pattern in the database, and if there is no match, they give a warning.
Active Vulnerability Scan
In this class of methods, errors are provoked by certain actions, both known and unknown (fuzzy systems).
After that, the base analyzes the response to these actions. This class of methods is characteristic of vulnerability scanners.
To interpret the results, both classes of methods are applicable:
- There is a variant of analyzing the answers on the basis (for example, characteristic answers to typical SQL injection patterns), or behavioral analysis (how the goal followed and how it responds to the following requests).
For example, sending an incorrectly composed IP packet should lead to the fall of the vulnerable server, after which it will stop responding. - There is the option of detecting anomalous activity.
For example, if after sending an ICMP packet of 777 bytes in size with 0xDEADBEEF, the network level of network activity increases dramatically, and then it drops - this is an anomaly (the normal situation is that the level of network activity does not change).
Pluses are obvious:
- Proactive intrusion detection.
Any attack may never happen, but the network will remain vulnerable to it.
This class of methods will reveal potential vulnerabilities. - Network verification is similar to an attacker, which increases the chances of detecting vulnerabilities.
Minuses:
- Additional network load.
- The ability to implement a successful attack during the scanning process, for example, the DoS of some services.
- As a rule, dependence on the base of attacks, which is becoming obsolete.
- Unobvious minus - the illusion of security. It may feel that "if the scanner did not find anything, then everything is ok."
At the same time, the risks of directed attacks still remain. This risk, perhaps to a lesser extent, is the case for other classes of methods.
Classes of data interpretation methods
It is possible to distinguish the following classes, each of which may include several methods:
- Methods for detecting known violations.
- Anomaly detection methods.
- Mixed method, which includes both of the above.
Detection of known violations
It comes down to finding signs of already known attacks.
Benefits:
- Methods are almost not subject to false positives.
- Methods implemented as a comparison with the sample, as a rule, are performed fairly quickly and do not require large expenditures of resources.
The disadvantage of these methods is the inability to detect unknown attacks.
In the classic version, the implementation assumes a comparison of package signatures with signatures in the database. Comparison can be, as exact, and with a template or on a regular expression.
Most well-known IDS use methods of this class.
This may also include fuzzy comparison methods:
- The trained perceptron.
- Methods for comparing with fuzzy logic.
Anomaly detection
The point is to record a pattern of normal network activity and respond to deviations from this pattern.
Possible implementation in the form of a database of signatures "normal" packets on the network
and statistical deviation detection system , when the analyzer searches for some rare actions or activities. Events in the system are studied statistically to find those of them, the manifestation of which looks abnormal.
The detector below will attempt to detect anomalies.
Results presentation methods
As the analysis of existing solutions has shown, two types of methods are commonly used:
- Two-grade. The system answers the question "Are there any problems?", The answer is "yes / no."
This method is typical for academic articles and research, but it can be applicable for writing modules or sensors of real SOW. - Multi-class The system answers the question "What are the problems now?". If there are no problems, then "no".
The result can also be presented with a certain level of confidence. And any neural network represents a result, not as "yes" or "no", but as a set of probabilities of results.
In the existing approaches, the probability is approximated to unity or zero (for example, from all possible attacks at the network output, the first one is chosen with the highest probability) and nowhere else is it taken into account.
IDS Types
At this stage, the parameter "Method of presenting the result" is of no interest (it was introduced to separate the detectors).
from itertools import product data_type_class = ('host_data', 'net_data', 'hybrid_data') analyzer_type_class = ('passive_analyzer', 'active_analyzer', 'mixed_analyzer') detector_type_class = ('known_issues_detector', 'anomaly_detector', 'mixed_detector') liter = list(product(data_type_class, analyzer_type_class, detector_type_class)) print(len(liter)) for n, i in enumerate(liter): print('Type {}:\ndata_type_class = {}, analyzer_type_class = {}, detector_type_class = {}.'.format(n, i[0], i[1], i[2])) print('-' * 10) Next, I will reduce them to fewer types:
- Types: 0, 1, 2. IDS node-level , based on the search for signatures, detecting anomalous behavior, or hybrid . There are a lot of similar IDS. It is also possible to include antiviruses.
- Types: 3, 4, 5. A node-level vulnerability scanner based on a signature search that detects anomalous behavior, or a hybrid one. Unclosed vulnerabilities scanners. The functionality is built into antiviruses. With a stretch as a representative of the group it is possible to allocate AVZ . Further I separately do not sort this type.
- Types: 6, 7, 8. Hybrid node-level IDS , both tracking attacks passively and allowing active scanning.
- Types: 9, 10, 11. IDS network level , based on the search for signatures, detecting anomalous behavior, or hybrid.
- Types: 12, 13, 14. Network level vulnerability scanner , based on signature search, detecting abnormal reactions, or hybrid.
- Types: 15, 16, 17. Hybrid IDS network level , detecting abnormal behavior, or hybrid. It should allow both scanning and detecting problems in passive mode. These are complex network level solutions . Usually, they do not exist in this form and merge with the following type.
- Type 26 (types 18-25 usually do not make sense separately and are reduced to this type). A hybrid system based on agents , each of which can have one of the above types.
- A separate item that does not fall into the classification, because it is not an IDS, but takes place as an auxiliary system, is a honeypot .
Below are examples of existing solutions that fall into the above groups.
Existing solutions
Operating systems
Snort
Snort is a classic network-level IDS and analyzes traffic for matching the rule base (in fact, with the signature base). That is, this system is looking for known violations. For Snort, it is possible to easily implement your module, which was done in one of the works. On the basis of Snort, many well-known commercial solutions have been implemented , including Russian ones .
In addition to working with the signature database, built on the basis of Snort IDS, it may well be composed of heuristic, neural network and similar detection modules. At a minimum, there is a working statistical anomaly detector for Snort.
Suricata
Suricata as well as Snort is a network level system.
This system has several features:
- Signature bases are Snort compatible.
- Evaluates not only the network / transport layer, but also works at the level of application protocols.
- It is possible to implement rules on Lua, an interpreted language, which expands the range of possibilities.
- It can analyze the traffic between two hosts, in general, and not just individual packets / connections. This allows, for example, to detect password guessing attempts.
- There is an IP reputation subsystem that allows you to assign a "reputation level" to each IP address.
That is, this system, although they detect known violations, as well as the previous one, has greater adaptability and the ability to learn (the reputation level of a host can change during the operation of the system and influence its decision making).
Bro
Platform for creating network-level IDS. It is a hybrid system, with a focus on the detection of known violations. Works at the transport, network and application levels. Supports your scripting language.
It is possible to detect anomalies, for example, multiple connections to services on different ports are not typical of normal node behavior, which will be detected.
This is implemented, firstly, on the basis of checks on the transmitted data for normality (for example, a TCP packet with all the flags set , probably something is wrong, despite the fact that it is correct).
Secondly, based on policies that describe how the network should function normally.
Bro not only detects attacks, but also helps in diagnosing network problems (claimed functionality).
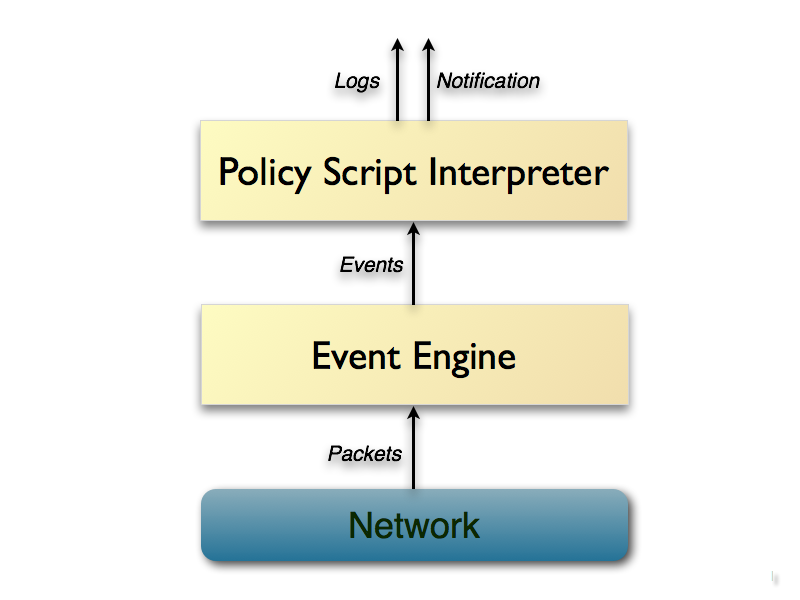
Technically, Bro is implemented quite interestingly: it does not analyze traffic directly by the signs, but drives packets on an “event machine,” which converts the stream of packets into a series of high-level events.
This machine can be considered an additional level of abstraction, which allows you to display network (usually, though not necessarily) activity in terms of policy neutrals (events simply signal that something has happened, but why the event engine doesn’t say anything, .k is the task of the policy interpreter).
For example, an HTTP request will be converted to an http_request event with the appropriate parameters, which is passed to the analysis level.
The policy interpreter executes the script in which event handlers are installed. These handlers can calculate statistical traffic parameters. At the same time, handlers can keep the context, and not just respond to a single package.
That is, the temporal dynamics, the “history” of the flow are taken into account.
A brief outline of the Bro core:
TripWire , OSSEC , Samhain
In classical terminology, these are representatives of host-level systems.
Combine methods for the detection of known violations and methods for finding abnormal activity.
The anomaly search mechanism is based on the fact that during installation, the system saves system hashes and meta information about them in the database. When upgrading operating system packages, hashes are recalculated.
In the case of an uncontrolled change of any observed file, after a while the system will let you know about it (as a rule, the scanner is started by the scheduler, although systems that react to FS events are possible).
In addition to monitoring files, these systems can monitor processes, connections, and analyze system logs.
Also, they can use the base of known violations, which is periodically updated.
Some have a centralized interface that allows you to analyze data simultaneously on many nodes.
Not bad the essence is described on the site Samhain:
This method provides a number of ways to ensure that your computer resides.
It can also be used as a standalone system.
OSSEC deployment example: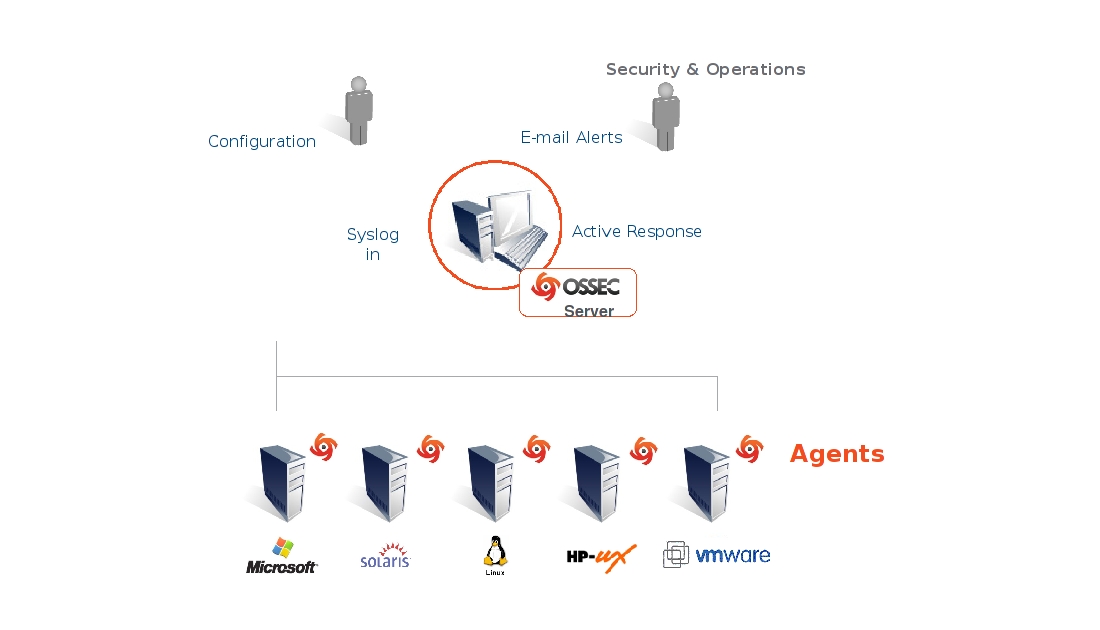
Prelude SIEM , OSSIM
These are hybrid systems positioned as SIEM . Prelude combines a sensor network and analyzer. It is stated that this system provides an increased level of security, since a hacker can bypass a single IDS, but the complexity of circumventing multiple protection mechanisms increases exponentially.
The system scales well: according to the developers, the sensor network can cover the continent or even the world.
The system is compatible with many existing IDS data format (IDMEF) including: AuditD, Nepenthes, NuFW, OSSEC, Pam, Samhain, Sancp, Snort, Suricata, Kismet, etc.
Approximately the same thing can be said about OSSIM.
Vulnerability Scanners
These are systems that are actively searching for vulnerabilities on a site or on a network.
The simplest are looking for only known problems that exist in the database; more serious systems can combine both methods for detecting known violations and abnormal activity.
Vulnerability scanners appeared a long time ago and they had quite a lot to make them.
I will cite only a few well-known products that are on the market:
- Nmap is a free network scanner, which is used rather as an auxiliary tool: it allows you to determine the OS, get a list of open ports and services that opened them, etc. ... At the same time, it includes the Netmap Scripting Engine, which allows you to build automated ones solutions (for example, to combine scanning with running brute force on the necessary ports).
- XSpider is a classic vulnerability scanner with a base, heuristics, search for anomalous behavior, etc.
- Metasploit is a framework containing exploit kits, scanners, ready loads launched on the attacked node, etc ... Allows you to conduct a study of a node or group of nodes on susceptibility to known vulnerabilities.
- OpenVAS is a framework that combines a scanner and a solution for managing vulnerabilities. It contains a database with more than 50,000 vulnerabilities, identifies the specified targets and checks their reaction to the use of the corresponding exploits.
The capabilities of these systems on the example of the above XSpider:
- Identification of services on random ports to identify server vulnerabilities with non-standard configuration.
- The heuristic method of determining the types and names of services (HTTP, FTP, SMTP, POP3, DNS, SSH, etc.) to determine the real name of the server and the correct operation of the checks.
- Handling RPC services (Windows and * nix) with full identification, including the definition of a detailed computer configuration.
- Check for weak password protection: optimized password selection in almost all services that require authentication.
- In-depth analysis of website content, including the identification of vulnerabilities in scripts: SQLi, XSS, launching arbitrary programs, etc.
- Analyzing the structure of HTTP servers to find weaknesses in the configuration.
- Extended check of nodes running Windows.
- Checks for non-standard DoS attacks.
- Ability to automate.
Honeypot
Systems specially made vulnerable in order to attract attacks to them. Can be used to investigate the actions of an attacker.
Deployment Example:
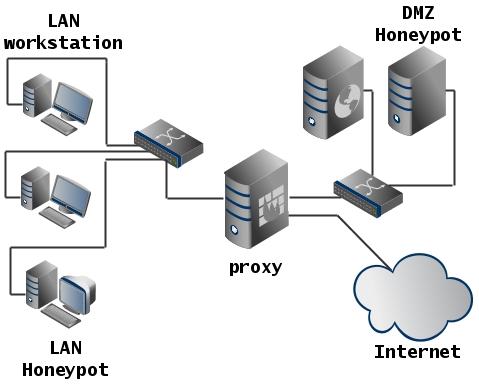
- HoneyWeb is a honeypot that launches WEB services.
- Honeyd - allows you to create virtual hosts that run services. Can emulate different OS.
In general, there are quite a few ready-made honeypot solutions.
This may also include tarpits , which slow down the attack (increases the requirements for the attacker's resources, gives time for response), which may be considered the “active” version of the honeypot.
Variants of approaches to solving the problem and neural network solutions
In [14], an analysis was made of references to the widely used KDD database.
- Support Vector Machines - 24.
- Decisive trees - 19.
- Genetic algorithms - 16.
- The principal component method is 13.
- Particle swarm method - 9.
- Search for k-nearest neighbors - 9.
- K-value clustering - 9.
- Naive Bayes classifiers - 9.
- Neural networks, multilayer perceptron - 8.
- Genetic programming - 6.
- Rough sets - 6.
- Bayesian networks - 5.
- A forest of random trees - 5.
- Artificial Immune System - 5.
- The use of fuzzy rules (Fuzzy Rules Mining) - 4.
- Neural networks (self-organizing maps) - 4.
As you can see, neural networks in the works for 2015-2016 are not so widely represented, moreover, as a rule, these are direct distribution networks.
In practice, the solutions, including those given above, are mainly based on the following analysis technologies:
- Methods for detecting known violations:
- Signature analysis. Comparison of data signatures, or behavior signatures with a signature in an updated database. A signature can also be represented by a pattern or regular expression.
- Expert systems based on rules [1].
- Methods for the detection of anomalies:
- Threshold detectors (responsive, for example, to a stable excess of CPU load on the server) [1].
- Statistical systems (for example, Bayesian classifiers , or systems of trained classifiers ).
- Behavioral analysis (possibly attributed to vague rules).
- Using the model "artificial immune system" .
About some solutions that I have not touched, it is also possible to look here .
These solutions have their own problems:
- The signature search does not respond to unknown attacks, and not large enough changes to the attack signature are enough for the detector to no longer detect it.
- For rule-based expert systems, as well as for signature search, it is required to keep the database up to date.
- For rule-based systems, minor variations in the sequence of actions during an attack can affect the activity-rule comparison process to such an extent that the attack is not detected by the appropriate detection mechanism. Increasing the level of distraction in such systems provides a partial solution to this problem, but this greatly increases the number of false positives [10].
- Rule-based systems often do not have sufficient flexibility in the structure of rules.
- Statistical systems are not sensitive to the order of events (which is not true for all existing systems).
- For them and for threshold detectors, it is difficult to set the threshold values of the characteristics monitored by the attack detection system.
- Over time, statistical systems can be retrained by intruders so that the attacking actions are treated as normal.
Neural networks, first of all, should be considered as a replacement for statistical detectors of anomalies, or additions to them, but to some extent, they can replace signature search and other methods.
In addition, hybrid systems are likely to be applied in the near future.
Neural networks have their advantages :
- The ability to analyze incomplete input data or a noisy signal.
- No need to formalize knowledge (replaced by learning).
- Fault tolerance: the failure of some network elements or the destruction of communication does not always make the network completely inoperable.
- The ability to easily parallelize work.
- Neural networks require less operator intervention.
- There is a chance of detecting unknown attacks.
- The network is able to learn automatically and in the process.
- The ability to process multidimensional data without a significant increase in labor intensity.
And disadvantages :
- Most approaches are heuristic and often do not lead to unambiguous solutions.
- To build a model of an object based on neurosis, preliminary network training is required, which requires computational and time costs.
- In order to train the network, it is necessary to prepare a training and test sample, which is not always easy.
- In some cases, network training leads to deadlocks: networks can be subject to retraining, or not converging.
- Network behavior may not always be unambiguously predictable, which introduces the risk of false positives or the omission of incidents.
- It is difficult to explain why the network made a decision (the problem of verbalization).
- Therefore, it is impossible to guarantee the repeatability and uniqueness of the results.
Existing solutions and proposals based on neural networks
Summarize the approaches described in the literature:
- In the works of Mustafayev [2], Zhigulin and Podvorchan [6], Halenar [7] proposed to use a multilayer perceptron, previously trained on the basis of attacks (for example, KDD).
- In the work of Haibo Zhang [3] and co-authors it is proposed to use a neural network and wavelet transform.
- Min-Joo Kang [4] and co-authors use deep learning to detect problems in the vehicle’s on-board CAN network.
- Talalaev and coauthors [8] propose to use a recirculating neural network or the principal component method for compressing the feature space, after which they investigate the use of both a double-layer perceptron and a Kohonen map.
- Balakhontsev and colleagues [10] use a three-layer perceptron.
- Kornev and Pylkin [13] investigate existing approaches and indicate the possibility of using perceptrons with different numbers of layers, single-layer classifiers for detecting the normal state, as well as a hybrid network consisting of a Kohonen map and a perceptron.
And practically used:
- DeepInstinct offers solutions using deep learning, but I have not found any details of the technology they use, and no comparisons. Judging by the fact that they have presented on the site, they use training with a teacher.
- According to unofficial data, the development of neural network detectors is actively conducted by companies, including several Russian ones.
Conclusions on existing solutions
SPD management and monitoring tools are evolving towards comprehensive solutions .
Modern systems, in general, strive to not only perform the narrow task of intrusion detection, but also help in diagnosing network faults, while implementing both anomaly detection methods and methods for detecting known violations.
The structure of such an integrated system includes various sensors distributed over the network, which can be both passive and active. Using sensor data, which can be whole IDS instances, the central correlator (in Prelude SIEM terminology) analyzes the overall network condition.
At the same time, the volume of data that needs to be processed and their dimension increase.
In the literature, mainly neural network methods based on perceptrons or Kohonen cards are considered. There is an unfilled niche in the form of developments, based on the latest research on competitive neural networks or convolutional networks. Although these methods are well proven in related areas where complex analysis is required.
There is a clear gap between the needs of the market and the offer in the form of existing solutions.
Therefore, it makes sense to conduct research in this direction.
Module design
Possible attacks
, , , [16].
- , , IDS ( , ARP , Ethernet, PPP , ).
, 90- — 2000-, .
Ping of the Death , IP Spoofing , SYN flood ARP cache poison , - DNS .
(, SQL injection PHP injection ), , XSS CSRF , , .
. ( , ).
Active Directory , "-" SMB .
HTTPS HSTS , SSL stripping .
, (, , HeartBleed ), .
, .
DoS , , .
(, SYN Flood) , , (, HTTP ) DDoS.
.
- , . , , DoS: , Windows- , SSH . , .
, "" (, , ftpd, sendmail ), . , : , pdf , ...
IDS : , , . , , , .
. , .
, : , , .
, , , .
- , (, : , — , ). ( , ). .
:
- .
- , , .
- , , .
, .
" ", ICMP ping. .
— TCP SYN .
, IDS . , .
, , :
- , . : .
- . , ( ).
.
:
- .
- .
- /.
- (, ).
- .
- ( , ).
- (, ).
, , , , ( : , , ).
. , , , CPU.
, .. . , , .
, - , , , , , ( "" ). , (, , ), IDS.
:
- : DoS/DDoS.
- .
- . , .
.
Scanning
:
- : n m . , , .
- : n m p .
IDS: , .
, .
, .
, , :
- CPU ().
- IO ( ).
- ( ).
- ( ).
.
[12] :
- TCP , , IP .
- ICMP , IP ,
ICMP_ID. - IP , UDP .
[6] , NSL-KDD .
duration— .protocol_type— : TCP, UDP ..service— : HTTP, FTP, TELNET ..flag— : .scr_bytes— .dst_bytes— .land— 1 / /.wrong_fragments— "" .urgent— (urgent) .hot— "" .num_failed_logins— .logged_in— 1 — , 0 .num_compromised— .root_shell— 1 — , 0 — .su_attempted— 1 — "su root", 0 — . .num_root— "root".num_file_creations— .num_shells— " ".num_access_files— .num_outbound_cmds— ftp .is_host_login— 1 — "host" .is_quest_login— 1 — "".count— 2 .srv_count— 2 ..serror_rate— syn .srv_serror_rate— syn .rerror_rate— rej .srv_rerror_rate— rej .same_srv_rate— .diff_srv_rate— .srv_diff_hast_rate— .dst_host_count— , .dst_host_srv_count— , .dst_host_same_srv_rate— , .dst_host_diff_srv_rate— , .dst_host_same_src_port_rate— .dst_host_srv_diff_host_rate— .dst_host_serror_rate— c syn -.dst_host_srv_serror_rate— c syn .dst_host_rerror_rate— c rej -.dst_host_srv_rerror_rate— c rej .
duration— ().protocol_type— (TCP, UDP, .).service— (HTTP, TELNET .).flag— c .src_bytes— .dst_bytes— .land— 1 ; 0 .wrong_fragment— "" .urgent— URG.
, .
, .
- ID protocol — , .
- Source port — TCP UDP.
- Destination port — TCP UDP.
- Source Address — IP .
- Destination Address — IP .
- ICMP type — ICMP .
- Length of data transferred — .
- FLAGS — .
- TCP window size — TCP .
, , , .
, NSL-KDD , . , . , , .
, [6] .
duration— .protocol_type— (TCP, UDP, .).service— (HTTP, TELNET .).flag— c .src_bytes— .dst_bytes— .land— 1 ; 0 .wrong_fragment— "" .urgent— URG.count— 2 ..srv_count— 2 ..serror_rate— syn .diff_srv_rate— .srv_diff_host_rate— .dst_host_srv_count— , .
IDS
IDS :
:
- .
- .
- .
:
- .
- .
- :
- .
- .
IDS, , . Prelude SIEM OSSIM, . , .
- – .
- – .
- – .
- – , .
.
. , , , .
, , .
, .
, , . , , , (, ), .
:
-> " ": " " -> " ": \n " " -> "": "" -> " ": \n " " -> : , : . . , IDS .
, , . , , , ( NSL-KDD). .
, ( , ), .
, , , .
, .
, (), , , , .
, :
, , , " ".
: , , .
- , , : .
( , , ).
, , , .
, ( NSL-KDD):

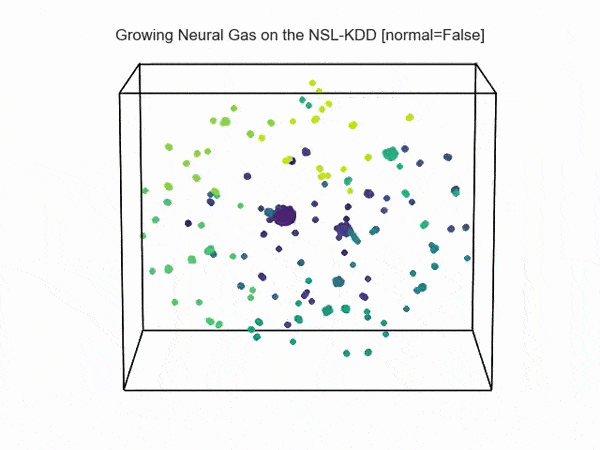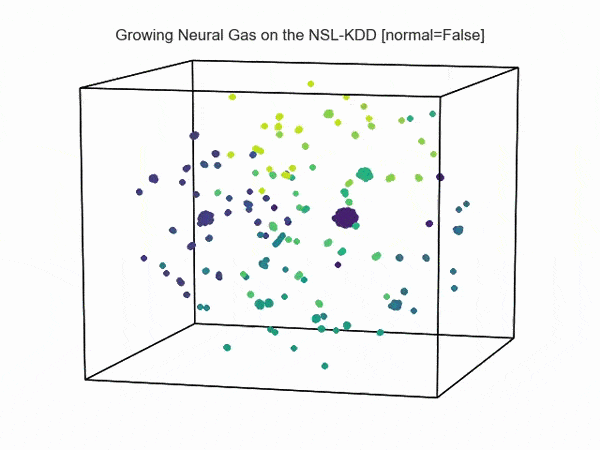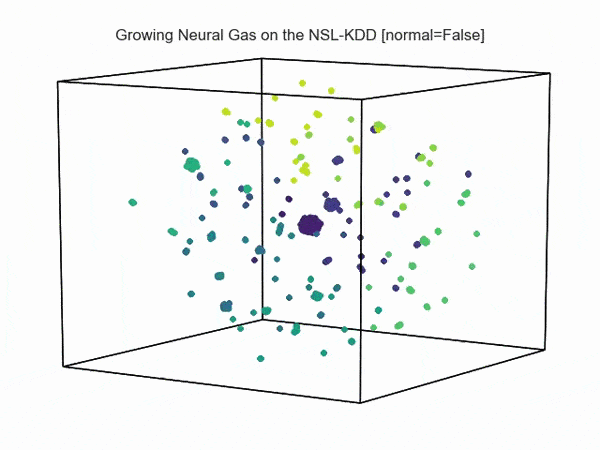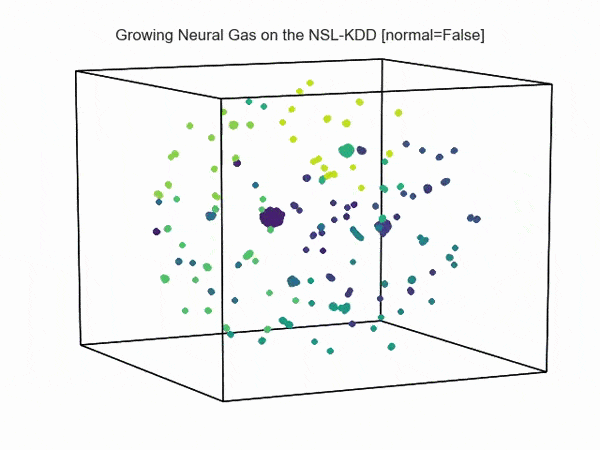
.., :
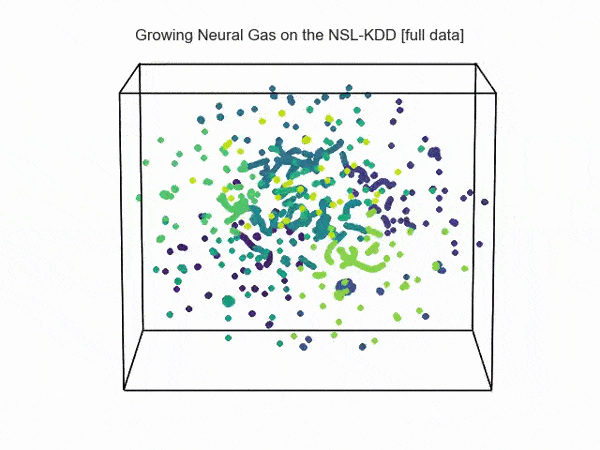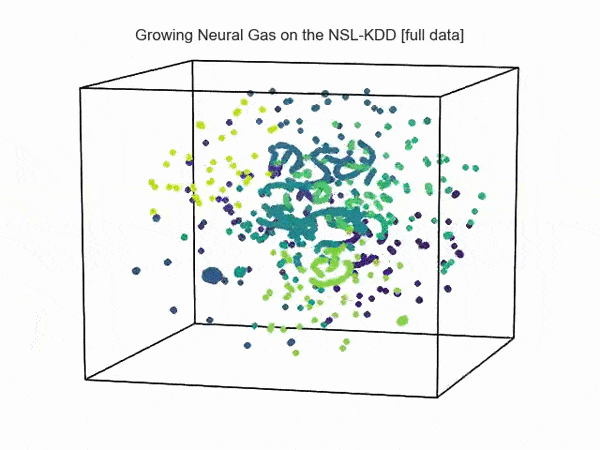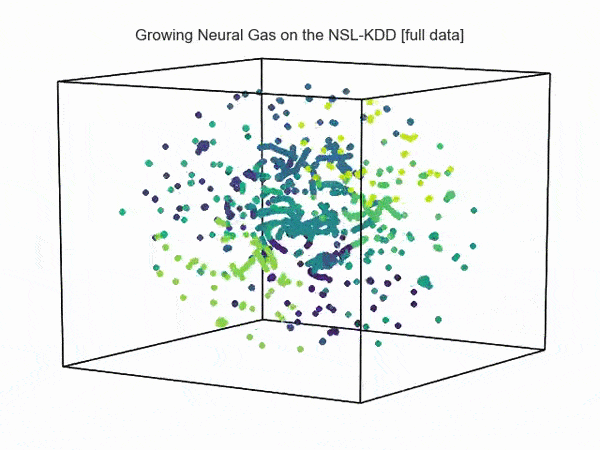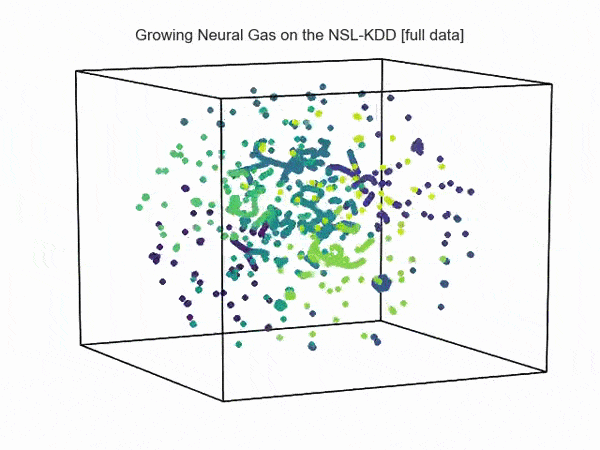
import hypertools as hyp from collections import OrderedDict import csv def read_ids_data(data_file, is_normal=True, labels_file='NSL_KDD/Field Names.csv', with_host=False): selected_parameters = ['duration', 'protocol_type', 'service', 'flag', 'src_bytes', 'dst_bytes', 'land', 'wrong_fragment', 'urgent'] # "Label" - "converter function" dictionary. label_dict = OrderedDict() result = [] with open(labels_file) as lf: labels = csv.reader(lf) for label in labels: if len(label) == 1 or label[1] == 'continuous': label_dict[label[0]] = lambda l: np.float64(l) elif label[1] == 'symbolic': label_dict[label[0]] = lambda l: sh(l) f_list = [i for i in label_dict.values()] n_list = [i for i in label_dict.keys()] data_type = lambda t: t == 'normal' if is_normal else t != 'normal' with open(data_file) as df: # data = csv.DictReader(df, label_dict.keys()) data = csv.reader(df) for d in data: if data_type(d[-2]): # Skip last two fields and add only specified fields. net_params = tuple(f_list[n](i) for n, i in enumerate(d[:-2]) if n_list[n] in selected_parameters) if with_host: host_params = generate_host_activity(is_normal) result.append(net_params + host_params) else: result.append(net_params) hyp.plot(np.array(result), '.', normalize='across', reduce='UMAP', ndims=3, n_clusters=10, animate='spin', palette='viridis', title='Growing Neural Gas on the NSL-KDD [normal={}]'.format(is_normal), # vectorizer='TfidfVectorizer', # precog=False, bullettime=True, chemtrails=True, tail_duration=100, duration=3, rotations=1, legend=False, explore=False, show=True, save_path='./video.mp4') read_ids_data('NSL_KDD/20 Percent Training Set.csv') read_ids_data('NSL_KDD/20 Percent Training Set.csv', is_normal=False) , " ".
, .
, , :
- .
- , .
, , .
: . , , , .
:
- . , .
- , .
, :
- .
- .
, .
: , .
. , (, , ).
. .
. , IDS.
, ( , 25%).
IDS . NSL-KDD.
:

@startuml start partition { : ; : \n ; } partition { : ; : ; if ( ) then () -[#blue]-> : ; if ( ) then () -[#green]-> : \n .; else () -[#blue]-> endif else () -[#green]-> : \n ; if ( ) then () -[#green]-> : ; else () -[#blue]-> : ; endif endif } stop @enduml , , .
, .
, , .
, .
, , 20 .
: . , .
— , .
, " " GNG , .
GNG , .
, :
- GNG [18] — , , [17], .
- IGNG [19] — . . . 20 " GNG", , ( [19]).
, , .
: , , .
, GNG ( , ), , .
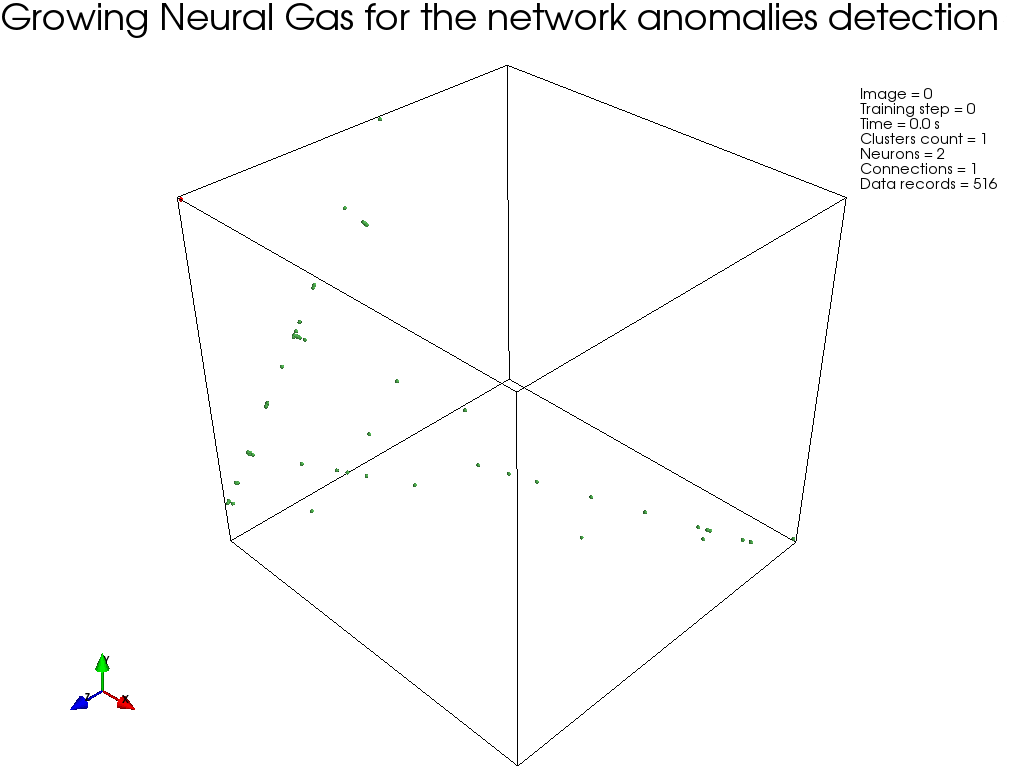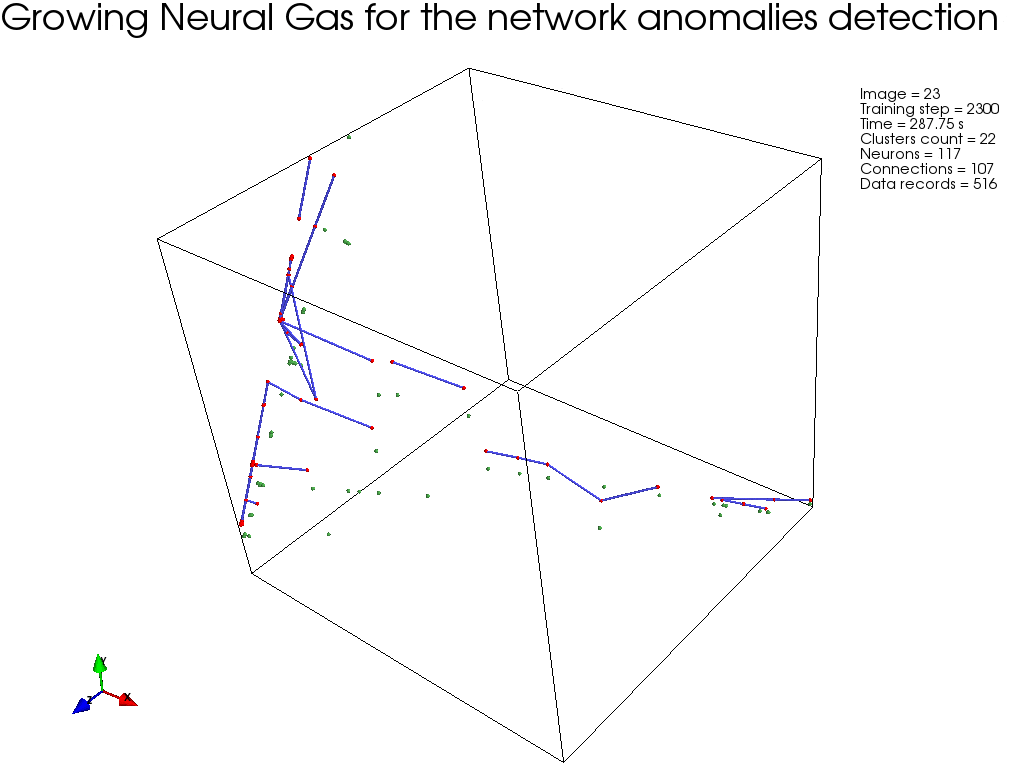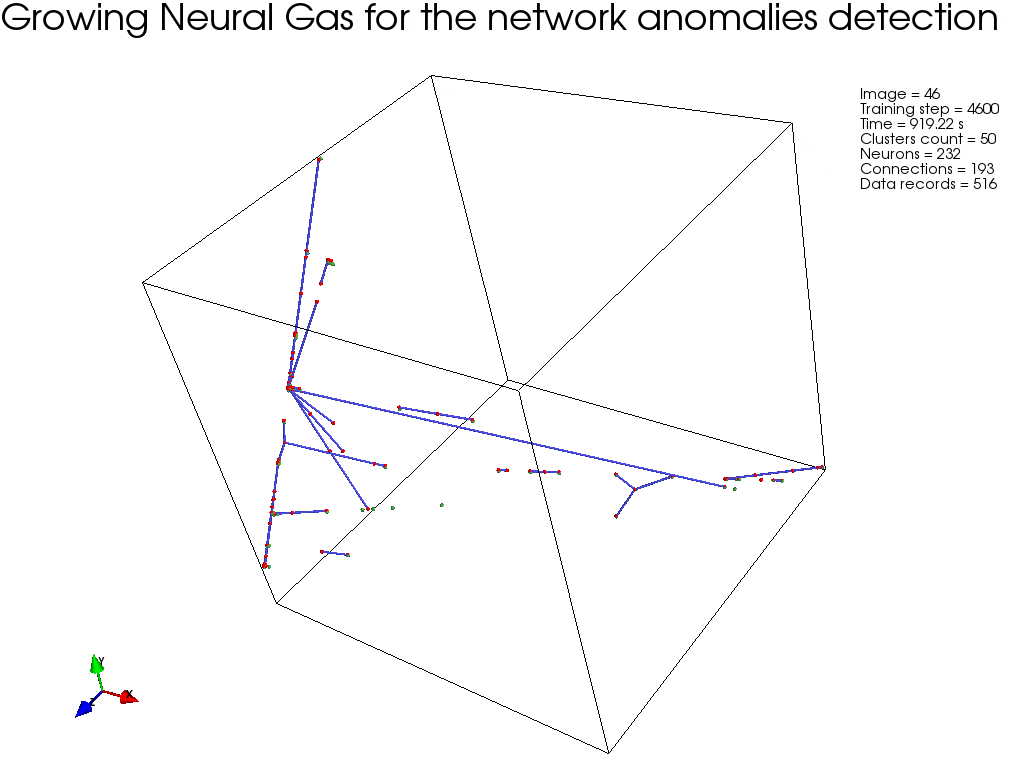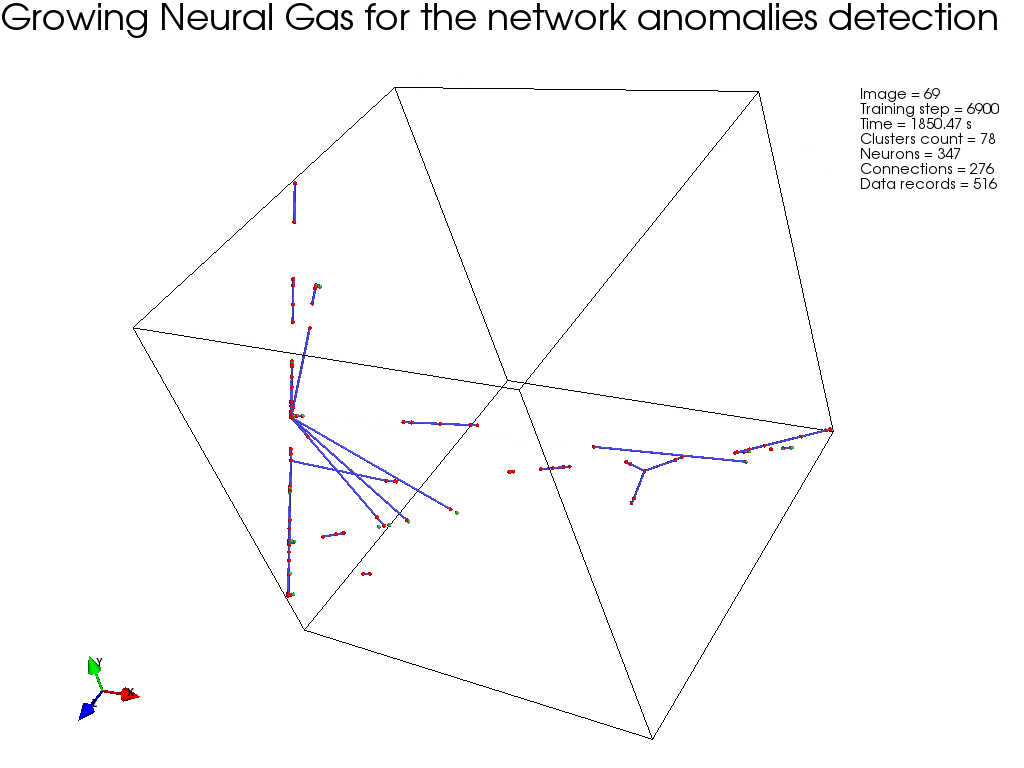
GNG, :
, Python. IGNG , , , .. . , IGNG .
main() , test_detector() , , .
test_detector() .
, , train() , , detect_anomalies() .
read_ids_data() .
NSL-KDD csv . : , , activity_type .
, , , generate_host_activity() , .
Numpy .
, , normalize() preprocessing Scikit-Learn . , .
, Graph NetworkX create_data_graph() .
.
. , .
GNG IGNG , . NeuralGas , .
train() __save_img() .
, .
, (, ) , , .
__save_img() draw_dots3d() , draw_graph3d() , : , .
Mayavi , — mayavi.points3d() .
, GIF, ImageIO .
:
https://github.com/artiomn/GNG
. , , , (. i train : , ). , , IGNG.
( , Github ) .
#!/usr/bin/env python # -*- coding: utf-8 -*- from abc import ABCMeta, abstractmethod from math import sqrt from mayavi import mlab import operator import imageio from collections import OrderedDict from scipy.spatial.distance import euclidean from sklearn import preprocessing import csv import numpy as np import networkx as nx import re import os import shutil import sys import glob from past.builtins import xrange from future.utils import iteritems import time def sh(s): sum = 0 for i, c in enumerate(s): sum += i * ord(c) return sum def create_data_graph(dots): """Create the graph and returns the networkx version of it 'G'.""" count = 0 G = nx.Graph() for i in dots: G.add_node(count, pos=(i)) count += 1 return G def get_ra(ra=0, ra_step=0.3): while True: if ra >= 360: ra = 0 else: ra += ra_step yield ra def shrink_to_3d(data): result = [] for i in data: depth = len(i) if depth <= 3: result.append(i) else: sm = np.sum([(n) * v for n, v in enumerate(i[2:])]) if sm == 0: sm = 1 r = np.array([i[0], i[1], i[2]]) r *= sm r /= np.sum(r) result.append(r) return preprocessing.normalize(result, axis=0, norm='max') def draw_dots3d(dots, edges, fignum, clear=True, title='', size=(1024, 768), graph_colormap='viridis', bgcolor=(1, 1, 1), node_color=(0.3, 0.65, 0.3), node_size=0.01, edge_color=(0.3, 0.3, 0.9), edge_size=0.003, text_size=0.14, text_color=(0, 0, 0), text_coords=[0.84, 0.75], text={}, title_size=0.3, angle=get_ra()): # https://stackoverflow.com/questions/17751552/drawing-multiplex-graphs-with-networkx # numpy array of x, y, z positions in sorted node order xyz = shrink_to_3d(dots) if fignum == 0: mlab.figure(fignum, bgcolor=bgcolor, fgcolor=text_color, size=size) # Mayavi is buggy, and following code causes sockets leak. #if mlab.options.offscreen: # mlab.figure(fignum, bgcolor=bgcolor, fgcolor=text_color, size=size) #elif fignum == 0: # mlab.figure(fignum, bgcolor=bgcolor, fgcolor=text_color, size=size) if clear: mlab.clf() # the x,y, and z co-ordinates are here # manipulate them to obtain the desired projection perspective pts = mlab.points3d(xyz[:, 0], xyz[:, 1], xyz[:, 2], scale_factor=node_size, scale_mode='none', color=node_color, #colormap=graph_colormap, resolution=20, transparent=False) mlab.text(text_coords[0], text_coords[1], '\n'.join(['{} = {}'.format(n, v) for n, v in text.items()]), width=text_size) if clear: mlab.title(title, height=0.95) mlab.roll(next(angle)) mlab.orientation_axes(pts) mlab.outline(pts) """ for i, (x, y, z) in enumerate(xyz): label = mlab.text(x, y, str(i), z=z, width=text_size, name=str(i), color=text_color) label.property.shadow = True """ pts.mlab_source.dataset.lines = edges tube = mlab.pipeline.tube(pts, tube_radius=edge_size) mlab.pipeline.surface(tube, color=edge_color) #mlab.show() # interactive window def draw_graph3d(graph, fignum, *args, **kwargs): graph_pos = nx.get_node_attributes(graph, 'pos') edges = np.array([e for e in graph.edges()]) dots = np.array([graph_pos[v] for v in sorted(graph)], dtype='float64') draw_dots3d(dots, edges, fignum, *args, **kwargs) def generate_host_activity(is_normal): # Host loads is changed only in 25% cases. attack_percent = 25 up_level = (20, 30) # CPU load in percent. cpu_load = (10, 30) # Disk IO per second. iops = (10, 50) # Memory consumption in percent. mem_cons = (30, 60) # Memory consumption in Mb/s. netw_act = (10, 50) cur_up_level = 0 if not is_normal and np.random.randint(0, 100) < attack_percent: cur_up_level = np.random.randint(*up_level) cpu_load = np.random.randint(cur_up_level + cpu_load[0], cur_up_level + cpu_load[1]) iops = np.random.randint(cur_up_level + iops[0], cur_up_level + iops[1]) mem_cons = np.random.randint(cur_up_level + mem_cons[0], cur_up_level + mem_cons[1]) netw_act = np.random.randint(cur_up_level + netw_act[0], cur_up_level + netw_act[1]) return cpu_load, iops, mem_cons, netw_act def read_ids_data(data_file, activity_type='normal', labels_file='NSL_KDD/Field Names.csv', with_host=False): selected_parameters = ['duration', 'protocol_type', 'service', 'flag', 'src_bytes', 'dst_bytes', 'land', 'wrong_fragment', 'urgent', 'serror_rate', 'diff_srv_rate', 'srv_diff_host_rate', 'dst_host_srv_count', 'count'] # "Label" - "converter function" dictionary. label_dict = OrderedDict() result = [] with open(labels_file) as lf: labels = csv.reader(lf) for label in labels: if len(label) == 1 or label[1] == 'continuous': label_dict[label[0]] = lambda l: np.float64(l) elif label[1] == 'symbolic': label_dict[label[0]] = lambda l: sh(l) f_list = [i for i in label_dict.values()] n_list = [i for i in label_dict.keys()] if activity_type == 'normal': data_type = lambda t: t == 'normal' elif activity_type == 'abnormal': data_type = lambda t: t != 'normal' elif activity_type == 'full': data_type = lambda t: True else: raise ValueError('`activity_type` must be "normal", "abnormal" or "full"') print('Reading {} activity from the file "{}" [generated host data {} included]...'. format(activity_type, data_file, 'was' if with_host else 'was not')) with open(data_file) as df: # data = csv.DictReader(df, label_dict.keys()) data = csv.reader(df) for d in data: if data_type(d[-2]): # Skip last two fields and add only specified fields. net_params = tuple(f_list[n](i) for n, i in enumerate(d[:-2]) if n_list[n] in selected_parameters) if with_host: host_params = generate_host_activity(activity_type != 'abnormal') result.append(net_params + host_params) else: result.append(net_params) print('Records count: {}'.format(len(result))) return result class NeuralGas(): __metaclass__ = ABCMeta def __init__(self, data, surface_graph=None, output_images_dir='images'): self._graph = nx.Graph() self._data = data self._surface_graph = surface_graph # Deviation parameters. self._dev_params = None self._output_images_dir = output_images_dir # Nodes count. self._count = 0 if os.path.isdir(output_images_dir): shutil.rmtree('{}'.format(output_images_dir)) print("Ouput images will be saved in: {0}".format(output_images_dir)) os.makedirs(output_images_dir) self._start_time = time.time() @abstractmethod def train(self, max_iterations=100, save_step=0): raise NotImplementedError() def number_of_clusters(self): return nx.number_connected_components(self._graph) def detect_anomalies(self, data, threshold=5, train=False, save_step=100): anomalies_counter, anomaly_records_counter, normal_records_counter = 0, 0, 0 anomaly_level = 0 start_time = self._start_time = time.time() for i, d in enumerate(data): risk_level = self.test_node(d, train) if risk_level != 0: anomaly_records_counter += 1 anomaly_level += risk_level if anomaly_level > threshold: anomalies_counter += 1 #print('Anomaly was detected [count = {}]!'.format(anomalies_counter)) anomaly_level = 0 else: normal_records_counter += 1 if i % save_step == 0: tm = time.time() - start_time print('Abnormal records = {}, Normal records = {}, Detection time = {} s, Time per record = {} s'. format(anomaly_records_counter, normal_records_counter, round(tm, 2), tm / i if i else 0)) tm = time.time() - start_time print('{} [abnormal records = {}, normal records = {}, detection time = {} s, time per record = {} s]'. format('Anomalies were detected (count = {})'.format(anomalies_counter) if anomalies_counter else 'Anomalies weren\'t detected', anomaly_records_counter, normal_records_counter, round(tm, 2), tm / len(data))) return anomalies_counter > 0 def test_node(self, node, train=False): n, dist = self._determine_closest_vertice(node) dev = self._calculate_deviation_params() dev = dev.get(frozenset(nx.node_connected_component(self._graph, n)), dist + 1) dist_sub_dev = dist - dev if dist_sub_dev > 0: return dist_sub_dev if train: self._dev_params = None self._train_on_data_item(node) return 0 @abstractmethod def _train_on_data_item(self, data_item): raise NotImplementedError() @abstractmethod def _save_img(self, fignum, training_step): """.""" raise NotImplementedError() def _calculate_deviation_params(self, distance_function_params={}): if self._dev_params is not None: return self._dev_params clusters = {} dcvd = self._determine_closest_vertice dlen = len(self._data) #dmean = np.mean(self._data, axis=1) #deviation = 0 for node in self._data: n = dcvd(node, **distance_function_params) cluster = clusters.setdefault(frozenset(nx.node_connected_component(self._graph, n[0])), [0, 0]) cluster[0] += n[1] cluster[1] += 1 clusters = {k: sqrt(v[0]/v[1]) for k, v in clusters.items()} self._dev_params = clusters return clusters def _determine_closest_vertice(self, curnode): """.""" pos = nx.get_node_attributes(self._graph, 'pos') kv = zip(*pos.items()) distances = np.linalg.norm(kv[1] - curnode, ord=2, axis=1) i0 = np.argsort(distances)[0] return kv[0][i0], distances[i0] def _determine_2closest_vertices(self, curnode): """Where this curnode is actually the x,y index of the data we want to analyze.""" pos = nx.get_node_attributes(self._graph, 'pos') l_pos = len(pos) if l_pos == 0: return None, None elif l_pos == 1: return pos[0], None kv = zip(*pos.items()) # Calculate Euclidean distance (2-norm of difference vectors) and get first two indexes of the sorted array. # Or a Euclidean-closest nodes index. distances = np.linalg.norm(kv[1] - curnode, ord=2, axis=1) i0, i1 = np.argsort(distances)[0:2] winner1 = tuple((kv[0][i0], distances[i0])) winner2 = tuple((kv[0][i1], distances[i1])) return winner1, winner2 class IGNG(NeuralGas): """Incremental Growing Neural Gas multidimensional implementation""" def __init__(self, data, surface_graph=None, eps_b=0.05, eps_n=0.0005, max_age=10, a_mature=1, output_images_dir='images'): """.""" NeuralGas.__init__(self, data, surface_graph, output_images_dir) self._eps_b = eps_b self._eps_n = eps_n self._max_age = max_age self._a_mature = a_mature self._num_of_input_signals = 0 self._fignum = 0 self._max_train_iters = 0 # Initial value is a standard deviation of the data. self._d = np.std(data) def train(self, max_iterations=100, save_step=0): """IGNG training method""" self._dev_params = None self._max_train_iters = max_iterations fignum = self._fignum self._save_img(fignum, 0) CHS = self.__calinski_harabaz_score igng = self.__igng data = self._data if save_step < 1: save_step = max_iterations old = 0 calin = CHS() i_count = 0 start_time = self._start_time = time.time() while old - calin <= 0: print('Iteration {0:d}...'.format(i_count)) i_count += 1 steps = 1 while steps <= max_iterations: for i, x in enumerate(data): igng(x) if i % save_step == 0: tm = time.time() - start_time print('Training time = {} s, Time per record = {} s, Training step = {}, Clusters count = {}, Neurons = {}, CHI = {}'. format(round(tm, 2), tm / (i if i and i_count == 0 else len(data)), i_count, self.number_of_clusters(), len(self._graph), old - calin) ) self._save_img(fignum, i_count) fignum += 1 steps += 1 self._d -= 0.1 * self._d old = calin calin = CHS() print('Training complete, clusters count = {}, training time = {} s'.format(self.number_of_clusters(), round(time.time() - start_time, 2))) self._fignum = fignum def _train_on_data_item(self, data_item): steps = 0 igng = self.__igng # while steps < self._max_train_iters: while steps < 5: igng(data_item) steps += 1 def __long_train_on_data_item(self, data_item): """.""" np.append(self._data, data_item) self._dev_params = None CHS = self.__calinski_harabaz_score igng = self.__igng data = self._data max_iterations = self._max_train_iters old = 0 calin = CHS() i_count = 0 # Strictly less. while old - calin < 0: print('Training with new normal node, step {0:d}...'.format(i_count)) i_count += 1 steps = 0 if i_count > 100: print('BUG', old, calin) break while steps < max_iterations: igng(data_item) steps += 1 self._d -= 0.1 * self._d old = calin calin = CHS() def _calculate_deviation_params(self, skip_embryo=True): return super(IGNG, self)._calculate_deviation_params(distance_function_params={'skip_embryo': skip_embryo}) def __calinski_harabaz_score(self, skip_embryo=True): graph = self._graph nodes = graph.nodes extra_disp, intra_disp = 0., 0. # CHI = [B / (c - 1)]/[W / (n - c)] # Total numb er of neurons. #ns = nx.get_node_attributes(self._graph, 'n_type') c = len([v for v in nodes.values() if v['n_type'] == 1]) if skip_embryo else len(nodes) # Total number of data. n = len(self._data) # Mean of the all data. mean = np.mean(self._data, axis=1) pos = nx.get_node_attributes(self._graph, 'pos') for node, k in pos.items(): if skip_embryo and nodes[node]['n_type'] == 0: # Skip embryo neurons. continue mean_k = np.mean(k) extra_disp += len(k) * np.sum((mean_k - mean) ** 2) intra_disp += np.sum((k - mean_k) ** 2) return (1. if intra_disp == 0. else extra_disp * (n - c) / (intra_disp * (c - 1.))) def _determine_closest_vertice(self, curnode, skip_embryo=True): """Where this curnode is actually the x,y index of the data we want to analyze.""" pos = nx.get_node_attributes(self._graph, 'pos') nodes = self._graph.nodes distance = sys.maxint for node, position in pos.items(): if skip_embryo and nodes[node]['n_type'] == 0: # Skip embryo neurons. continue dist = euclidean(curnode, position) if dist < distance: distance = dist return node, distance def __get_specific_nodes(self, n_type): return [n for n, p in nx.get_node_attributes(self._graph, 'n_type').items() if p == n_type] def __igng(self, cur_node): """Main IGNG training subroutine""" # find nearest unit and second nearest unit winner1, winner2 = self._determine_2closest_vertices(cur_node) graph = self._graph nodes = graph.nodes d = self._d # Second list element is a distance. if winner1 is None or winner1[1] >= d: # 0 - is an embryo type. graph.add_node(self._count, pos=cur_node, n_type=0, age=0) winner_node1 = self._count self._count += 1 return else: winner_node1 = winner1[0] # Second list element is a distance. if winner2 is None or winner2[1] >= d: # 0 - is an embryo type. graph.add_node(self._count, pos=cur_node, n_type=0, age=0) winner_node2 = self._count self._count += 1 graph.add_edge(winner_node1, winner_node2, age=0) return else: winner_node2 = winner2[0] # Increment the age of all edges, emanating from the winner. for e in graph.edges(winner_node1, data=True): e[2]['age'] += 1 w_node = nodes[winner_node1] # Move the winner node towards current node. w_node['pos'] += self._eps_b * (cur_node - w_node['pos']) neighbors = nx.all_neighbors(graph, winner_node1) a_mature = self._a_mature for n in neighbors: c_node = nodes[n] # Move all direct neighbors of the winner. c_node['pos'] += self._eps_n * (cur_node - c_node['pos']) # Increment the age of all direct neighbors of the winner. c_node['age'] += 1 if c_node['n_type'] == 0 and c_node['age'] >= a_mature: # Now, it's a mature neuron. c_node['n_type'] = 1 # Create connection with age == 0 between two winners. graph.add_edge(winner_node1, winner_node2, age=0) max_age = self._max_age # If there are ages more than maximum allowed age, remove them. age_of_edges = nx.get_edge_attributes(graph, 'age') for edge, age in iteritems(age_of_edges): if age >= max_age: graph.remove_edge(edge[0], edge[1]) # If it causes isolated vertix, remove that vertex as well. #graph.remove_nodes_from(nx.isolates(graph)) for node, v in nodes.items(): if v['n_type'] == 0: # Skip embryo neurons. continue if not graph.neighbors(node): graph.remove_node(node) def _save_img(self, fignum, training_step): """.""" title='Incremental Growing Neural Gas for the network anomalies detection' if self._surface_graph is not None: text = OrderedDict([ ('Image', fignum), ('Training step', training_step), ('Time', '{} s'.format(round(time.time() - self._start_time, 2))), ('Clusters count', self.number_of_clusters()), ('Neurons', len(self._graph)), (' Mature', len(self.__get_specific_nodes(1))), (' Embryo', len(self.__get_specific_nodes(0))), ('Connections', len(self._graph.edges)), ('Data records', len(self._data)) ]) draw_graph3d(self._surface_graph, fignum, title=title) graph = self._graph if len(graph) > 0: #graph_pos = nx.get_node_attributes(graph, 'pos') #nodes = sorted(self.get_specific_nodes(1)) #dots = np.array([graph_pos[v] for v in nodes], dtype='float64') #edges = np.array([e for e in graph.edges(nodes) if e[0] in nodes and e[1] in nodes]) #draw_dots3d(dots, edges, fignum, clear=False, node_color=(1, 0, 0)) draw_graph3d(graph, fignum, clear=False, node_color=(1, 0, 0), title=title, text=text) mlab.savefig("{0}/{1}.png".format(self._output_images_dir, str(fignum))) #mlab.close(fignum) class GNG(NeuralGas): """Growing Neural Gas multidimensional implementation""" def __init__(self, data, surface_graph=None, eps_b=0.05, eps_n=0.0006, max_age=15, lambda_=20, alpha=0.5, d=0.005, max_nodes=1000, output_images_dir='images'): """.""" NeuralGas.__init__(self, data, surface_graph, output_images_dir) self._eps_b = eps_b self._eps_n = eps_n self._max_age = max_age self._lambda = lambda_ self._alpha = alpha self._d = d self._max_nodes = max_nodes self._fignum = 0 self.__add_initial_nodes() def train(self, max_iterations=10000, save_step=50, stop_on_chi=False): """.""" self._dev_params = None self._save_img(self._fignum, 0) graph = self._graph max_nodes = self._max_nodes d = self._d ld = self._lambda alpha = self._alpha update_winner = self.__update_winner data = self._data CHS = self.__calinski_harabaz_score old = 0 calin = CHS() start_time = self._start_time = time.time() train_step = self.__train_step for i in xrange(1, max_iterations): tm = time.time() - start_time print('Training time = {} s, Time per record = {} s, Training step = {}/{}, Clusters count = {}, Neurons = {}'. format(round(tm, 2), tm / len(data), i, max_iterations, self.number_of_clusters(), len(self._graph)) ) for x in data: update_winner(x) train_step(i, alpha, ld, d, max_nodes, True, save_step, graph, update_winner) old = calin calin = CHS() # Stop on the enough clusterization quality. if stop_on_chi and old - calin > 0: break print('Training complete, clusters count = {}, training time = {} s'.format(self.number_of_clusters(), round(time.time() - start_time, 2))) def __train_step(self, i, alpha, ld, d, max_nodes, save_img, save_step, graph, update_winner): g_nodes = graph.nodes # Step 8: if number of input signals generated so far if i % ld == 0 and len(graph) < max_nodes: # Find a node with the largest error. errorvectors = nx.get_node_attributes(graph, 'error') node_largest_error = max(errorvectors.items(), key=operator.itemgetter(1))[0] # Find a node from neighbor of the node just found, with a largest error. neighbors = graph.neighbors(node_largest_error) max_error_neighbor = None max_error = -1 for n in neighbors: ce = g_nodes[n]['error'] if ce > max_error: max_error = ce max_error_neighbor = n # Decrease error variable of other two nodes by multiplying with alpha. new_max_error = alpha * errorvectors[node_largest_error] graph.nodes[node_largest_error]['error'] = new_max_error graph.nodes[max_error_neighbor]['error'] = alpha * max_error # Insert a new unit half way between these two. self._count += 1 new_node = self._count graph.add_node(new_node, pos=self.__get_average_dist(g_nodes[node_largest_error]['pos'], g_nodes[max_error_neighbor]['pos']), error=new_max_error) # Insert edges between new node and other two nodes. graph.add_edge(new_node, max_error_neighbor, age=0) graph.add_edge(new_node, node_largest_error, age=0) # Remove edge between old nodes. graph.remove_edge(max_error_neighbor, node_largest_error) if True and i % save_step == 0: self._fignum += 1 self._save_img(self._fignum, i) # step 9: Decrease all error variables. for n in graph.nodes(): oe = g_nodes[n]['error'] g_nodes[n]['error'] -= d * oe def _train_on_data_item(self, data_item): """IGNG training method""" np.append(self._data, data_item) graph = self._graph max_nodes = self._max_nodes d = self._d ld = self._lambda alpha = self._alpha update_winner = self.__update_winner data = self._data train_step = self.__train_step #for i in xrange(1, 5): update_winner(data_item) train_step(0, alpha, ld, d, max_nodes, False, -1, graph, update_winner) def _calculate_deviation_params(self): return super(GNG, self)._calculate_deviation_params() def __add_initial_nodes(self): """Initialize here""" node1 = self._data[np.random.randint(0, len(self._data))] node2 = self._data[np.random.randint(0, len(self._data))] # make sure you dont select same positions if self.__is_nodes_equal(node1, node2): raise ValueError("Rerun ---------------> similar nodes selected") self._count = 0 self._graph.add_node(self._count, pos=node1, error=0) self._count += 1 self._graph.add_node(self._count, pos=node2, error=0) self._graph.add_edge(self._count - 1, self._count, age=0) def __is_nodes_equal(self, n1, n2): return len(set(n1) & set(n2)) == len(n1) def __update_winner(self, curnode): """.""" # find nearest unit and second nearest unit winner1, winner2 = self._determine_2closest_vertices(curnode) winner_node1 = winner1[0] winner_node2 = winner2[0] win_dist_from_node = winner1[1] graph = self._graph g_nodes = graph.nodes # Update the winner error. g_nodes[winner_node1]['error'] += + win_dist_from_node**2 # Move the winner node towards current node. g_nodes[winner_node1]['pos'] += self._eps_b * (curnode - g_nodes[winner_node1]['pos']) eps_n = self._eps_n # Now update all the neighbors distances. for n in nx.all_neighbors(graph, winner_node1): g_nodes[n]['pos'] += eps_n * (curnode - g_nodes[n]['pos']) # Update age of the edges, emanating from the winner. for e in graph.edges(winner_node1, data=True): e[2]['age'] += 1 # Create or zeroe edge between two winner nodes. graph.add_edge(winner_node1, winner_node2, age=0) # if there are ages more than maximum allowed age, remove them age_of_edges = nx.get_edge_attributes(graph, 'age') max_age = self._max_age for edge, age in age_of_edges.items(): if age >= max_age: graph.remove_edge(edge[0], edge[1]) # If it causes isolated vertix, remove that vertex as well. for node in g_nodes: if not graph.neighbors(node): graph.remove_node(node) def __get_average_dist(self, a, b): """.""" return (a + b) / 2 def __calinski_harabaz_score(self): graph = self._graph nodes = graph.nodes extra_disp, intra_disp = 0., 0. # CHI = [B / (c - 1)]/[W / (n - c)] # Total numb er of neurons. #ns = nx.get_node_attributes(self._graph, 'n_type') c = len(nodes) # Total number of data. n = len(self._data) # Mean of the all data. mean = np.mean(self._data, axis=1) pos = nx.get_node_attributes(self._graph, 'pos') for node, k in pos.items(): mean_k = np.mean(k) extra_disp += len(k) * np.sum((mean_k - mean) ** 2) intra_disp += np.sum((k - mean_k) ** 2) def _save_img(self, fignum, training_step): """.""" title = 'Growing Neural Gas for the network anomalies detection' if self._surface_graph is not None: text = OrderedDict([ ('Image', fignum), ('Training step', training_step), ('Time', '{} s'.format(round(time.time() - self._start_time, 2))), ('Clusters count', self.number_of_clusters()), ('Neurons', len(self._graph)), ('Connections', len(self._graph.edges)), ('Data records', len(self._data)) ]) draw_graph3d(self._surface_graph, fignum, title=title) graph = self._graph if len(graph) > 0: draw_graph3d(graph, fignum, clear=False, node_color=(1, 0, 0), title=title, text=text) mlab.savefig("{0}/{1}.png".format(self._output_images_dir, str(fignum))) def sort_nicely(limages): """Numeric string sort""" def convert(text): return int(text) if text.isdigit() else text def alphanum_key(key): return [convert(c) for c in re.split('([0-9]+)', key)] limages = sorted(limages, key=alphanum_key) return limages def convert_images_to_gif(output_images_dir, output_gif): """Convert a list of images to a gif.""" image_dir = "{0}/*.png".format(output_images_dir) list_images = glob.glob(image_dir) file_names = sort_nicely(list_images) images = [imageio.imread(fn) for fn in file_names] imageio.mimsave(output_gif, images) def test_detector(use_hosts_data, max_iters, alg, output_images_dir='images', output_gif='output.gif'): """Detector quality testing routine""" #data = read_ids_data('NSL_KDD/20 Percent Training Set.csv') frame = '-' * 70 training_set = 'NSL_KDD/Small Training Set.csv' #training_set = 'NSL_KDD/KDDTest-21.txt' testing_set = 'NSL_KDD/KDDTest-21.txt' #testing_set = 'NSL_KDD/KDDTrain+.txt' print('{}\n{}\n{}'.format(frame, '{} detector training...'.format(alg.__name__), frame)) data = read_ids_data(training_set, activity_type='normal', with_host=use_hosts_data) data = preprocessing.normalize(np.array(data, dtype='float64'), axis=1, norm='l1', copy=False) G = create_data_graph(data) gng = alg(data, surface_graph=G, output_images_dir=output_images_dir) gng.train(max_iterations=max_iters, save_step=50) print('Saving GIF file...') convert_images_to_gif(output_images_dir, output_gif) print('{}\n{}\n{}'.format(frame, 'Applying detector to the normal activity using the training set...', frame)) gng.detect_anomalies(data) for a_type in ['abnormal', 'full']: print('{}\n{}\n{}'.format(frame, 'Applying detector to the {} activity using the training set...'.format(a_type), frame)) d_data = read_ids_data(training_set, activity_type=a_type, with_host=use_hosts_data) d_data = preprocessing.normalize(np.array(d_data, dtype='float64'), axis=1, norm='l1', copy=False) gng.detect_anomalies(d_data) dt = OrderedDict([('normal', None), ('abnormal', None), ('full', None)]) for a_type in dt.keys(): print('{}\n{}\n{}'.format(frame, 'Applying detector to the {} activity using the testing set without adaptive learning...'.format(a_type), frame)) d = read_ids_data(testing_set, activity_type=a_type, with_host=use_hosts_data) dt[a_type] = d = preprocessing.normalize(np.array(d, dtype='float64'), axis=1, norm='l1', copy=False) gng.detect_anomalies(d, save_step=1000, train=False) for a_type in ['full']: print('{}\n{}\n{}'.format(frame, 'Applying detector to the {} activity using the testing set with adaptive learning...'.format(a_type), frame)) gng.detect_anomalies(dt[a_type], train=True, save_step=1000) def main(): """Entry point""" start_time = time.time() mlab.options.offscreen = True test_detector(use_hosts_data=False, max_iters=7000, alg=GNG, output_gif='gng_wohosts.gif') print('Working time = {}'.format(round(time.time() - start_time, 2))) test_detector(use_hosts_data=True, max_iters=7000, alg=GNG, output_gif='gng_whosts.gif') print('Working time = {}'.format(round(time.time() - start_time, 2))) test_detector(use_hosts_data=False, max_iters=100, alg=IGNG, output_gif='igng_wohosts.gif') print('Working time = {}'.format(round(time.time() - start_time, 2))) test_detector(use_hosts_data=True, max_iters=100, alg=IGNG, output_gif='igng_whosts.gif') print('Full working time = {}'.format(round(time.time() - start_time, 2))) return 0 if __name__ == "__main__": exit(main()) results
. , ( ), .
, , .
, .
, , .
, .
:
- GNG: 7000.
- : 516.
- : 495.
- : 2152.
- : 9698.
- : 11850.
---------------------------------------------------------------------- GNG detector training... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was not included]... Records count: 516 Ouput images will be saved in: images Training time = 0.0 s, Time per record = 5.54461811864e-09 s, Training step = 1/7000, Clusters count = 1, Neurons = 2 Training time = 0.04 s, Time per record = 7.09027282951e-05 s, Training step = 2/7000, Clusters count = 1, Neurons = 2 Training time = 0.07 s, Time per record = 0.000142523022585 s, Training step = 3/7000, Clusters count = 1, Neurons = 2 Training time = 0.11 s, Time per record = 0.000216007694718 s, Training step = 4/7000, Clusters count = 1, Neurons = 2 Training time = 0.16 s, Time per record = 0.000302943610406 s, Training step = 5/7000, Clusters count = 1, Neurons = 2 Training time = 0.2 s, Time per record = 0.000389085259548 s, Training step = 6/7000, Clusters count = 1, Neurons = 2 Training time = 0.25 s, Time per record = 0.000486224658729 s, Training step = 7/7000, Clusters count = 1, Neurons = 2 Training time = 0.3 s, Time per record = 0.000579897285432 s, Training step = 8/7000, Clusters count = 1, Neurons = 2 Training time = 0.35 s, Time per record = 0.000673654929612 s, Training step = 9/7000, Clusters count = 1, Neurons = 2 ... Training time = 1889.7 s, Time per record = 3.66220002119 s, Training step = 6986/7000, Clusters count = 78, Neurons = 351 Training time = 1890.16 s, Time per record = 3.66309242239 s, Training step = 6987/7000, Clusters count = 78, Neurons = 351 Training time = 1890.61 s, Time per record = 3.6639701858 s, Training step = 6988/7000, Clusters count = 78, Neurons = 351 Training time = 1891.07 s, Time per record = 3.66486349586 s, Training step = 6989/7000, Clusters count = 78, Neurons = 351 Training time = 1891.52 s, Time per record = 3.66574243797 s, Training step = 6990/7000, Clusters count = 78, Neurons = 351 Training time = 1891.98 s, Time per record = 3.66663252291 s, Training step = 6991/7000, Clusters count = 78, Neurons = 351 Training time = 1892.43 s, Time per record = 3.6675082556 s, Training step = 6992/7000, Clusters count = 78, Neurons = 351 Training time = 1892.9 s, Time per record = 3.66840752705 s, Training step = 6993/7000, Clusters count = 78, Neurons = 351 Training time = 1893.35 s, Time per record = 3.66929203087 s, Training step = 6994/7000, Clusters count = 78, Neurons = 351 Training time = 1893.81 s, Time per record = 3.6701756531 s, Training step = 6995/7000, Clusters count = 78, Neurons = 351 Training time = 1894.26 s, Time per record = 3.67105510068 s, Training step = 6996/7000, Clusters count = 78, Neurons = 351 Training time = 1894.71 s, Time per record = 3.67192640508 s, Training step = 6997/7000, Clusters count = 78, Neurons = 351 Training time = 1895.18 s, Time per record = 3.67282555408 s, Training step = 6998/7000, Clusters count = 78, Neurons = 351 Training time = 1895.63 s, Time per record = 3.67370033726 s, Training step = 6999/7000, Clusters count = 78, Neurons = 351 Training complete, clusters count = 78, training time = 1896.09 s Saving GIF file... ---------------------------------------------------------------------- Applying detector to the normal activity using the training set... ---------------------------------------------------------------------- Abnormal records = 0, Normal records = 1, Detection time = 0.2 s, Time per record = 0 s Abnormal records = 0, Normal records = 101, Detection time = 0.24 s, Time per record = 0.00244196891785 s Abnormal records = 0, Normal records = 201, Detection time = 0.28 s, Time per record = 0.00141963005066 s Abnormal records = 0, Normal records = 301, Detection time = 0.32 s, Time per record = 0.00107904990514 s Abnormal records = 0, Normal records = 401, Detection time = 0.36 s, Time per record = 0.000907952189445 s Abnormal records = 0, Normal records = 501, Detection time = 0.4 s, Time per record = 0.000804873943329 s Anomalies weren't detected [abnormal records = 0, normal records = 516, detection time = 0.41 s, time per record = 0.000791241956312 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the training set... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was not included]... Records count: 495 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 62, Normal records = 39, Detection time = 0.04 s, Time per record = 0.000383739471436 s Abnormal records = 133, Normal records = 68, Detection time = 0.08 s, Time per record = 0.000377835035324 s Abnormal records = 198, Normal records = 103, Detection time = 0.12 s, Time per record = 0.000388882954915 s Abnormal records = 269, Normal records = 132, Detection time = 0.15 s, Time per record = 0.000385674834251 s Anomalies were detected (count = 7) [abnormal records = 344, normal records = 151, detection time = 0.19 s, time per record = 0.000390153461032 s] ---------------------------------------------------------------------- Applying detector to the full activity using the training set... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was not included]... Records count: 1011 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 29, Normal records = 72, Detection time = 0.04 s, Time per record = 0.000397310256958 s Abnormal records = 58, Normal records = 143, Detection time = 0.08 s, Time per record = 0.000394929647446 s Abnormal records = 90, Normal records = 211, Detection time = 0.12 s, Time per record = 0.000392502943675 s Abnormal records = 123, Normal records = 278, Detection time = 0.16 s, Time per record = 0.000393797159195 s Abnormal records = 156, Normal records = 345, Detection time = 0.2 s, Time per record = 0.000392875671387 s Abnormal records = 188, Normal records = 413, Detection time = 0.24 s, Time per record = 0.000391929944356 s Abnormal records = 218, Normal records = 483, Detection time = 0.27 s, Time per record = 0.000391151223864 s Abnormal records = 259, Normal records = 542, Detection time = 0.31 s, Time per record = 0.000390258729458 s Abnormal records = 294, Normal records = 607, Detection time = 0.35 s, Time per record = 0.000389169851939 s Abnormal records = 335, Normal records = 666, Detection time = 0.39 s, Time per record = 0.000388996839523 s Anomalies were detected (count = 7) [abnormal records = 344, normal records = 667, detection time = 0.39 s, time per record = 0.000388572524964 s] ---------------------------------------------------------------------- Applying detector to the normal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was not included]... Records count: 2152 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 368, Normal records = 633, Detection time = 0.4 s, Time per record = 0.000396910905838 s Abnormal records = 737, Normal records = 1264, Detection time = 0.81 s, Time per record = 0.000405857920647 s Anomalies were detected (count = 7) [abnormal records = 794, normal records = 1358, detection time = 0.88 s, time per record = 0.00040672259703 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was not included]... Records count: 9698 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 776, Normal records = 225, Detection time = 0.39 s, Time per record = 0.000390892028809 s Abnormal records = 1547, Normal records = 454, Detection time = 0.78 s, Time per record = 0.000388429045677 s Abnormal records = 2332, Normal records = 669, Detection time = 1.16 s, Time per record = 0.000386790037155 s Abnormal records = 3117, Normal records = 884, Detection time = 1.56 s, Time per record = 0.000389873743057 s Abnormal records = 3899, Normal records = 1102, Detection time = 1.95 s, Time per record = 0.000389337825775 s Abnormal records = 4700, Normal records = 1301, Detection time = 2.33 s, Time per record = 0.000388201673826 s Abnormal records = 5502, Normal records = 1499, Detection time = 2.71 s, Time per record = 0.000387295722961 s Abnormal records = 6277, Normal records = 1724, Detection time = 3.1 s, Time per record = 0.000387670874596 s Abnormal records = 7063, Normal records = 1938, Detection time = 3.49 s, Time per record = 0.000387644237942 s Anomalies were detected (count = 154) [abnormal records = 7605, normal records = 2093, detection time = 3.75 s, time per record = 0.000387096114935 s] ---------------------------------------------------------------------- Applying detector to the full activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was not included]... Records count: 11850 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 698, Normal records = 303, Detection time = 0.39 s, Time per record = 0.000389461040497 s Abnormal records = 1396, Normal records = 605, Detection time = 0.77 s, Time per record = 0.000386721491814 s Abnormal records = 2092, Normal records = 909, Detection time = 1.17 s, Time per record = 0.000389091014862 s Abnormal records = 2808, Normal records = 1193, Detection time = 1.56 s, Time per record = 0.00038931697607 s Abnormal records = 3519, Normal records = 1482, Detection time = 1.95 s, Time per record = 0.000389059782028 s Abnormal records = 4229, Normal records = 1772, Detection time = 2.33 s, Time per record = 0.000388749321302 s Abnormal records = 4957, Normal records = 2044, Detection time = 2.72 s, Time per record = 0.000388323715755 s Abnormal records = 5668, Normal records = 2333, Detection time = 3.11 s, Time per record = 0.00038857537508 s Abnormal records = 6376, Normal records = 2625, Detection time = 3.51 s, Time per record = 0.000389481120639 s Abnormal records = 7079, Normal records = 2922, Detection time = 3.89 s, Time per record = 0.000389060306549 s Abnormal records = 7800, Normal records = 3201, Detection time = 4.27 s, Time per record = 0.000388331630013 s Anomalies were detected (count = 161) [abnormal records = 8399, normal records = 3451, detection time = 4.61 s, time per record = 0.000388800504338 s] Working time = 1914.96 ---------------------------------------------------------------------- GNG detector training... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was included]... Records count: 516 Ouput images will be saved in: images Training time = 0.0 s, Time per record = 6.00666962853e-09 s, Training step = 1/7000, Clusters count = 1, Neurons = 2 Training time = 0.04 s, Time per record = 7.04014024069e-05 s, Training step = 2/7000, Clusters count = 1, Neurons = 2 Training time = 0.07 s, Time per record = 0.000143418940463 s, Training step = 3/7000, Clusters count = 1, Neurons = 2 Training time = 0.11 s, Time per record = 0.000213542649912 s, Training step = 4/7000, Clusters count = 1, Neurons = 2 Training time = 0.15 s, Time per record = 0.000284942083581 s, Training step = 5/7000, Clusters count = 1, Neurons = 2 Training time = 0.19 s, Time per record = 0.00037281781204 s, Training step = 6/7000, Clusters count = 1, Neurons = 2 Training time = 0.23 s, Time per record = 0.0004449033922 s, Training step = 7/7000, Clusters count = 1, Neurons = 2 Training time = 0.27 s, Time per record = 0.000518457372059 s, Training step = 8/7000, Clusters count = 1, Neurons = 2 Training time = 0.3 s, Time per record = 0.00058810701666 s, Training step = 9/7000, Clusters count = 1, Neurons = 2 Training time = 0.34 s, Time per record = 0.000659637672957 s, Training step = 10/7000, Clusters count = 1, Neurons = 2 Training time = 0.38 s, Time per record = 0.000728998073312 s, Training step = 11/7000, Clusters count = 1, Neurons = 2 Training time = 0.41 s, Time per record = 0.0007999751919 s, Training step = 12/7000, Clusters count = 1, Neurons = 2 ... Training time = 1832.99 s, Time per record = 3.55230925804 s, Training step = 6987/7000, Clusters count = 82, Neurons = 351 Training time = 1833.46 s, Time per record = 3.55321220973 s, Training step = 6988/7000, Clusters count = 82, Neurons = 351 Training time = 1833.92 s, Time per record = 3.5541091127 s, Training step = 6989/7000, Clusters count = 82, Neurons = 351 Training time = 1834.39 s, Time per record = 3.55502607194 s, Training step = 6990/7000, Clusters count = 82, Neurons = 351 Training time = 1834.85 s, Time per record = 3.55591803882 s, Training step = 6991/7000, Clusters count = 82, Neurons = 351 Training time = 1835.32 s, Time per record = 3.55682536844 s, Training step = 6992/7000, Clusters count = 82, Neurons = 351 Training time = 1835.78 s, Time per record = 3.55772192857 s, Training step = 6993/7000, Clusters count = 82, Neurons = 351 Training time = 1836.26 s, Time per record = 3.55863503107 s, Training step = 6994/7000, Clusters count = 82, Neurons = 351 Training time = 1836.72 s, Time per record = 3.55953075026 s, Training step = 6995/7000, Clusters count = 82, Neurons = 351 Training time = 1837.19 s, Time per record = 3.56043755823 s, Training step = 6996/7000, Clusters count = 82, Neurons = 351 Training time = 1837.65 s, Time per record = 3.56133750012 s, Training step = 6997/7000, Clusters count = 82, Neurons = 351 Training time = 1838.11 s, Time per record = 3.56223588766 s, Training step = 6998/7000, Clusters count = 82, Neurons = 351 Training time = 1838.58 s, Time per record = 3.56314611851 s, Training step = 6999/7000, Clusters count = 82, Neurons = 351 Training complete, clusters count = 82, training time = 1839.05 s Saving GIF file... ---------------------------------------------------------------------- Applying detector to the normal activity using the training set... ---------------------------------------------------------------------- Abnormal records = 0, Normal records = 1, Detection time = 0.22 s, Time per record = 0 s Abnormal records = 0, Normal records = 101, Detection time = 0.26 s, Time per record = 0.00264456033707 s Abnormal records = 0, Normal records = 201, Detection time = 0.31 s, Time per record = 0.00152621984482 s Abnormal records = 0, Normal records = 301, Detection time = 0.35 s, Time per record = 0.00115152041117 s Abnormal records = 0, Normal records = 401, Detection time = 0.39 s, Time per record = 0.000965242385864 s Abnormal records = 0, Normal records = 501, Detection time = 0.43 s, Time per record = 0.000851732254028 s Anomalies weren't detected [abnormal records = 0, normal records = 516, detection time = 0.43 s, time per record = 0.00083744387294 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the training set... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was included]... Records count: 495 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 45, Normal records = 56, Detection time = 0.04 s, Time per record = 0.000407500267029 s Abnormal records = 96, Normal records = 105, Detection time = 0.08 s, Time per record = 0.000401464700699 s Abnormal records = 151, Normal records = 150, Detection time = 0.12 s, Time per record = 0.000398120085398 s Abnormal records = 203, Normal records = 198, Detection time = 0.16 s, Time per record = 0.000406047701836 s Anomalies were detected (count = 5) [abnormal records = 265, normal records = 230, detection time = 0.2 s, time per record = 0.000403349808972 s] ---------------------------------------------------------------------- Applying detector to the full activity using the training set... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was included]... Records count: 1011 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 20, Normal records = 81, Detection time = 0.04 s, Time per record = 0.00041876077652 s Abnormal records = 50, Normal records = 151, Detection time = 0.08 s, Time per record = 0.000415489673615 s Abnormal records = 76, Normal records = 225, Detection time = 0.13 s, Time per record = 0.000419800281525 s Abnormal records = 101, Normal records = 300, Detection time = 0.17 s, Time per record = 0.000416232347488 s Abnormal records = 125, Normal records = 376, Detection time = 0.21 s, Time per record = 0.000413020133972 s Abnormal records = 155, Normal records = 446, Detection time = 0.25 s, Time per record = 0.000409803390503 s Abnormal records = 190, Normal records = 511, Detection time = 0.29 s, Time per record = 0.000409148420606 s Abnormal records = 220, Normal records = 581, Detection time = 0.33 s, Time per record = 0.000412831306458 s Abnormal records = 253, Normal records = 648, Detection time = 0.37 s, Time per record = 0.000414616531796 s Abnormal records = 289, Normal records = 712, Detection time = 0.41 s, Time per record = 0.000414277076721 s Anomalies were detected (count = 5) [abnormal records = 298, normal records = 713, detection time = 0.42 s, time per record = 0.000413806219129 s] ---------------------------------------------------------------------- Applying detector to the normal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was included]... Records count: 2152 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 324, Normal records = 677, Detection time = 0.4 s, Time per record = 0.000400429010391 s Abnormal records = 646, Normal records = 1355, Detection time = 0.8 s, Time per record = 0.000398404955864 s Anomalies were detected (count = 6) [abnormal records = 695, normal records = 1457, detection time = 0.86 s, time per record = 0.000401378675021 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was included]... Records count: 9698 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 716, Normal records = 285, Detection time = 0.39 s, Time per record = 0.000391721010208 s Abnormal records = 1424, Normal records = 577, Detection time = 0.79 s, Time per record = 0.000395292520523 s Abnormal records = 2144, Normal records = 857, Detection time = 1.18 s, Time per record = 0.000393549998601 s Abnormal records = 2877, Normal records = 1124, Detection time = 1.57 s, Time per record = 0.000393224000931 s Abnormal records = 3591, Normal records = 1410, Detection time = 1.97 s, Time per record = 0.000394224214554 s Abnormal records = 4337, Normal records = 1664, Detection time = 2.36 s, Time per record = 0.000393829345703 s Abnormal records = 5089, Normal records = 1912, Detection time = 2.77 s, Time per record = 0.000395649433136 s Abnormal records = 5798, Normal records = 2203, Detection time = 3.16 s, Time per record = 0.000395138502121 s Abnormal records = 6532, Normal records = 2469, Detection time = 3.55 s, Time per record = 0.000394880347782 s Anomalies were detected (count = 131) [abnormal records = 7027, normal records = 2671, detection time = 3.84 s, time per record = 0.000396096625262 s] ---------------------------------------------------------------------- Applying detector to the full activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was included]... Records count: 11850 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 622, Normal records = 379, Detection time = 0.4 s, Time per record = 0.000395487070084 s Abnormal records = 1251, Normal records = 750, Detection time = 0.79 s, Time per record = 0.000395890474319 s Abnormal records = 1876, Normal records = 1125, Detection time = 1.19 s, Time per record = 0.000395860671997 s Abnormal records = 2528, Normal records = 1473, Detection time = 1.6 s, Time per record = 0.000399308979511 s Abnormal records = 3184, Normal records = 1817, Detection time = 1.99 s, Time per record = 0.000397848176956 s Abnormal records = 3824, Normal records = 2177, Detection time = 2.39 s, Time per record = 0.000397575179736 s Abnormal records = 4498, Normal records = 2503, Detection time = 2.79 s, Time per record = 0.000398371560233 s Abnormal records = 5145, Normal records = 2856, Detection time = 3.18 s, Time per record = 0.00039776262641 s Abnormal records = 5792, Normal records = 3209, Detection time = 3.59 s, Time per record = 0.000398459778892 s Abnormal records = 6424, Normal records = 3577, Detection time = 3.98 s, Time per record = 0.000397949695587 s Abnormal records = 7080, Normal records = 3921, Detection time = 4.38 s, Time per record = 0.000397751092911 s Anomalies were detected (count = 138) [abnormal records = 7619, normal records = 4231, detection time = 4.72 s, time per record = 0.000398021931387 s] Working time = 3772.93 ---------------------------------------------------------------------- IGNG detector training... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was not included]... Records count: 516 Ouput images will be saved in: images Iteration 0... Training time = 0.0 s, Time per record = 4.85154085381e-08 s, Training step = 1, Clusters count = 1, Neurons = 1, CHI = -1.0 Training time = 0.48 s, Time per record = 0.000924686128779 s, Training step = 1, Clusters count = 11, Neurons = 22, CHI = -1.0 Training time = 0.96 s, Time per record = 0.00185829262401 s, Training step = 1, Clusters count = 10, Neurons = 28, CHI = -1.0 Training time = 1.45 s, Time per record = 0.00280647129976 s, Training step = 1, Clusters count = 11, Neurons = 31, CHI = -1.0 Training time = 1.93 s, Time per record = 0.0037418566933 s, Training step = 1, Clusters count = 11, Neurons = 33, CHI = -1.0 Training time = 2.42 s, Time per record = 0.00468139177145 s, Training step = 1, Clusters count = 11, Neurons = 36, CHI = -1.0 Training time = 2.9 s, Time per record = 0.00562962635543 s, Training step = 1, Clusters count = 14, Neurons = 40, CHI = -1.0 ... Training time = 542.57 s, Time per record = 1.05149506783 s, Training step = 1, Clusters count = 20, Neurons = 65, CHI = -1.0 Training time = 543.07 s, Time per record = 1.05246509692 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 543.57 s, Time per record = 1.05343477717 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 544.07 s, Time per record = 1.05440813395 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 544.59 s, Time per record = 1.05540252148 s, Training step = 1, Clusters count = 19, Neurons = 65, CHI = -1.0 Training time = 545.08 s, Time per record = 1.05635387398 s, Training step = 1, Clusters count = 20, Neurons = 65, CHI = -1.0 Training time = 545.58 s, Time per record = 1.05732676179 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 546.22 s, Time per record = 1.05855851395 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 546.72 s, Time per record = 1.0595340641 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 547.22 s, Time per record = 1.06049638571 s, Training step = 1, Clusters count = 20, Neurons = 65, CHI = -1.0 Training time = 547.71 s, Time per record = 1.06145610689 s, Training step = 1, Clusters count = 20, Neurons = 65, CHI = -1.0 Training time = 548.21 s, Time per record = 1.06243183955 s, Training step = 1, Clusters count = 20, Neurons = 65, CHI = -1.0 Training time = 548.7 s, Time per record = 1.06338078292 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 549.2 s, Time per record = 1.0643380252 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 549.7 s, Time per record = 1.06530855654 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 550.19 s, Time per record = 1.0662655932 s, Training step = 1, Clusters count = 19, Neurons = 65, CHI = -1.0 Training time = 550.69 s, Time per record = 1.06722681606 s, Training step = 1, Clusters count = 20, Neurons = 65, CHI = -1.0 Training time = 551.19 s, Time per record = 1.06818968427 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training complete, clusters count = 21, training time = 551.68 s Saving GIF file... ---------------------------------------------------------------------- Applying detector to the normal activity using the training set... ---------------------------------------------------------------------- Abnormal records = 0, Normal records = 1, Detection time = 0.53 s, Time per record = 0 s Abnormal records = 0, Normal records = 101, Detection time = 0.63 s, Time per record = 0.00633862018585 s Abnormal records = 0, Normal records = 201, Detection time = 0.73 s, Time per record = 0.00366881489754 s Abnormal records = 0, Normal records = 301, Detection time = 0.83 s, Time per record = 0.00277556975683 s Abnormal records = 0, Normal records = 401, Detection time = 0.93 s, Time per record = 0.00232824504375 s Abnormal records = 0, Normal records = 501, Detection time = 1.03 s, Time per record = 0.00206651210785 s Anomalies weren't detected [abnormal records = 0, normal records = 516, detection time = 1.05 s, time per record = 0.00203575598177 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the training set... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was not included]... Records count: 495 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 19, Normal records = 82, Detection time = 0.11 s, Time per record = 0.00108736038208 s Abnormal records = 41, Normal records = 160, Detection time = 0.21 s, Time per record = 0.00104064464569 s Abnormal records = 59, Normal records = 242, Detection time = 0.31 s, Time per record = 0.00102363348007 s Abnormal records = 78, Normal records = 323, Detection time = 0.41 s, Time per record = 0.00101397752762 s Anomalies were detected (count = 1) [abnormal records = 110, normal records = 385, detection time = 0.5 s, time per record = 0.00100804242221 s] ---------------------------------------------------------------------- Applying detector to the full activity using the training set... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was not included]... Records count: 1011 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 6, Normal records = 95, Detection time = 0.1 s, Time per record = 0.00101613998413 s Abnormal records = 18, Normal records = 183, Detection time = 0.2 s, Time per record = 0.00100832462311 s Abnormal records = 30, Normal records = 271, Detection time = 0.3 s, Time per record = 0.00100745360057 s Abnormal records = 38, Normal records = 363, Detection time = 0.41 s, Time per record = 0.00101710498333 s Abnormal records = 48, Normal records = 453, Detection time = 0.51 s, Time per record = 0.00101808595657 s Abnormal records = 58, Normal records = 543, Detection time = 0.61 s, Time per record = 0.00101492325465 s Abnormal records = 68, Normal records = 633, Detection time = 0.71 s, Time per record = 0.00101190873555 s Abnormal records = 76, Normal records = 725, Detection time = 0.81 s, Time per record = 0.00101043373346 s Abnormal records = 88, Normal records = 813, Detection time = 0.91 s, Time per record = 0.00100836992264 s Abnormal records = 105, Normal records = 896, Detection time = 1.01 s, Time per record = 0.00100904512405 s Anomalies were detected (count = 1) [abnormal records = 110, normal records = 901, detection time = 1.02 s, time per record = 0.00100898412521 s] ---------------------------------------------------------------------- Applying detector to the normal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was not included]... Records count: 2152 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 165, Normal records = 836, Detection time = 1.01 s, Time per record = 0.00101356911659 s Abnormal records = 344, Normal records = 1657, Detection time = 2.04 s, Time per record = 0.00101889550686 s Anomalies weren't detected [abnormal records = 372, normal records = 1780, detection time = 2.19 s, time per record = 0.00101830303447 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was not included]... Records count: 9698 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 289, Normal records = 712, Detection time = 1.01 s, Time per record = 0.0010106010437 s Abnormal records = 531, Normal records = 1470, Detection time = 2.03 s, Time per record = 0.00101473748684 s Abnormal records = 806, Normal records = 2195, Detection time = 3.05 s, Time per record = 0.00101690498988 s Abnormal records = 1085, Normal records = 2916, Detection time = 4.07 s, Time per record = 0.00101814824343 s Abnormal records = 1340, Normal records = 3661, Detection time = 5.1 s, Time per record = 0.00102022538185 s Abnormal records = 1615, Normal records = 4386, Detection time = 6.11 s, Time per record = 0.00101814981302 s Abnormal records = 1910, Normal records = 5091, Detection time = 7.12 s, Time per record = 0.00101696658134 s Abnormal records = 2177, Normal records = 5824, Detection time = 8.13 s, Time per record = 0.00101655423641 s Abnormal records = 2437, Normal records = 6564, Detection time = 9.14 s, Time per record = 0.00101562854979 s Anomalies were detected (count = 27) [abnormal records = 2602, normal records = 7096, detection time = 9.84 s, time per record = 0.00101498390724 s] ---------------------------------------------------------------------- Applying detector to the full activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was not included]... Records count: 11850 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 264, Normal records = 737, Detection time = 1.01 s, Time per record = 0.00101314496994 s Abnormal records = 496, Normal records = 1505, Detection time = 2.04 s, Time per record = 0.00101831400394 s Abnormal records = 748, Normal records = 2253, Detection time = 3.07 s, Time per record = 0.0010246480306 s Abnormal records = 1000, Normal records = 3001, Detection time = 4.08 s, Time per record = 0.00101987397671 s Abnormal records = 1264, Normal records = 3737, Detection time = 5.09 s, Time per record = 0.00101791639328 s Abnormal records = 1488, Normal records = 4513, Detection time = 6.1 s, Time per record = 0.00101627349854 s Abnormal records = 1761, Normal records = 5240, Detection time = 7.11 s, Time per record = 0.00101523229054 s Abnormal records = 2019, Normal records = 5982, Detection time = 8.11 s, Time per record = 0.00101378926635 s Abnormal records = 2282, Normal records = 6719, Detection time = 9.11 s, Time per record = 0.00101193467776 s Abnormal records = 2530, Normal records = 7471, Detection time = 10.11 s, Time per record = 0.00101094179153 s Abnormal records = 2781, Normal records = 8220, Detection time = 11.12 s, Time per record = 0.00101091491092 s Anomalies were detected (count = 28) [abnormal records = 2974, normal records = 8876, detection time = 11.97 s, time per record = 0.00101001697251 s] Working time = 5537.67 ---------------------------------------------------------------------- IGNG detector training... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was included]... Records count: 516 Ouput images will be saved in: images Iteration 0... Training time = 0.0 s, Time per record = 5.03636145777e-08 s, Training step = 1, Clusters count = 1, Neurons = 1, CHI = -1.0 Training time = 0.5 s, Time per record = 0.000959633856781 s, Training step = 1, Clusters count = 11, Neurons = 24, CHI = -1.0 Training time = 1.0 s, Time per record = 0.00193044566369 s, Training step = 1, Clusters count = 11, Neurons = 32, CHI = -1.0 Training time = 1.5 s, Time per record = 0.00290305069251 s, Training step = 1, Clusters count = 13, Neurons = 38, CHI = -1.0 Training time = 2.0 s, Time per record = 0.0038706590963 s, Training step = 1, Clusters count = 16, Neurons = 40, CHI = -1.0 Training time = 2.5 s, Time per record = 0.00484266669251 s, Training step = 1, Clusters count = 14, Neurons = 42, CHI = -1.0 Training time = 3.0 s, Time per record = 0.00581014156342 s, Training step = 1, Clusters count = 17, Neurons = 46, CHI = -1.0 ... Training time = 555.39 s, Time per record = 1.07633200958 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 555.9 s, Time per record = 1.07732224233 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 556.41 s, Time per record = 1.07831773093 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 556.93 s, Time per record = 1.07931286651 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 557.44 s, Time per record = 1.08031143421 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 557.95 s, Time per record = 1.08129599806 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 558.47 s, Time per record = 1.08229697305 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 558.98 s, Time per record = 1.08329910687 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 559.51 s, Time per record = 1.08431409496 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 560.02 s, Time per record = 1.08530911826 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 560.53 s, Time per record = 1.0863001707 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 561.05 s, Time per record = 1.08729686062 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 561.57 s, Time per record = 1.08831085854 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 562.07 s, Time per record = 1.08928472682 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 562.58 s, Time per record = 1.09027189525 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 563.09 s, Time per record = 1.09126116693 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 563.61 s, Time per record = 1.09226539523 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 564.13 s, Time per record = 1.09326583933 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 564.64 s, Time per record = 1.09426376385 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 565.15 s, Time per record = 1.09525539755 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 565.67 s, Time per record = 1.09626085259 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 566.2 s, Time per record = 1.09728375193 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 566.72 s, Time per record = 1.09829355996 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 567.24 s, Time per record = 1.09930341697 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training complete, clusters count = 22, training time = 567.74 s Saving GIF file... ---------------------------------------------------------------------- Applying detector to the normal activity using the training set... ---------------------------------------------------------------------- Abnormal records = 0, Normal records = 1, Detection time = 0.58 s, Time per record = 0 s Abnormal records = 0, Normal records = 101, Detection time = 0.69 s, Time per record = 0.00693108081818 s Abnormal records = 0, Normal records = 201, Detection time = 0.81 s, Time per record = 0.00403473496437 s Abnormal records = 0, Normal records = 301, Detection time = 0.92 s, Time per record = 0.00306363026301 s Abnormal records = 0, Normal records = 401, Detection time = 1.03 s, Time per record = 0.00257846474648 s Abnormal records = 0, Normal records = 501, Detection time = 1.15 s, Time per record = 0.00229543209076 s Anomalies weren't detected [abnormal records = 0, normal records = 516, detection time = 1.16 s, time per record = 0.00225632181463 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the training set... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was included]... Records count: 495 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 19, Normal records = 82, Detection time = 0.11 s, Time per record = 0.00114132165909 s Abnormal records = 42, Normal records = 159, Detection time = 0.23 s, Time per record = 0.00113487005234 s Abnormal records = 59, Normal records = 242, Detection time = 0.34 s, Time per record = 0.00112538019816 s Abnormal records = 79, Normal records = 322, Detection time = 0.45 s, Time per record = 0.00113292753696 s Anomalies were detected (count = 1) [abnormal records = 111, normal records = 384, detection time = 0.56 s, time per record = 0.0011294605756 s] ---------------------------------------------------------------------- Applying detector to the full activity using the training set... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was included]... Records count: 1011 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 6, Normal records = 95, Detection time = 0.11 s, Time per record = 0.00112770080566 s Abnormal records = 18, Normal records = 183, Detection time = 0.23 s, Time per record = 0.00115751504898 s Abnormal records = 30, Normal records = 271, Detection time = 0.34 s, Time per record = 0.00114720344543 s Abnormal records = 38, Normal records = 363, Detection time = 0.45 s, Time per record = 0.00113685250282 s Abnormal records = 47, Normal records = 454, Detection time = 0.57 s, Time per record = 0.00113049221039 s Abnormal records = 57, Normal records = 544, Detection time = 0.68 s, Time per record = 0.00112626830737 s Abnormal records = 67, Normal records = 634, Detection time = 0.79 s, Time per record = 0.00113069295883 s Abnormal records = 75, Normal records = 726, Detection time = 0.9 s, Time per record = 0.00112736135721 s Abnormal records = 87, Normal records = 814, Detection time = 1.01 s, Time per record = 0.00112713442908 s Abnormal records = 104, Normal records = 897, Detection time = 1.13 s, Time per record = 0.00112596797943 s Anomalies were detected (count = 1) [abnormal records = 109, normal records = 902, detection time = 1.14 s, time per record = 0.0011247270303 s] ---------------------------------------------------------------------- Applying detector to the normal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was included]... Records count: 2152 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 165, Normal records = 836, Detection time = 1.13 s, Time per record = 0.00113072776794 s Abnormal records = 343, Normal records = 1658, Detection time = 2.25 s, Time per record = 0.00112573599815 s Anomalies weren't detected [abnormal records = 371, normal records = 1781, detection time = 2.42 s, time per record = 0.00112456171929 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was included]... Records count: 9698 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 281, Normal records = 720, Detection time = 1.14 s, Time per record = 0.0011370780468 s Abnormal records = 514, Normal records = 1487, Detection time = 2.27 s, Time per record = 0.00113358747959 s Abnormal records = 785, Normal records = 2216, Detection time = 3.39 s, Time per record = 0.00112900535266 s Abnormal records = 1057, Normal records = 2944, Detection time = 4.51 s, Time per record = 0.00112772023678 s Abnormal records = 1297, Normal records = 3704, Detection time = 5.63 s, Time per record = 0.00112658400536 s Abnormal records = 1568, Normal records = 4433, Detection time = 6.76 s, Time per record = 0.00112644219398 s Abnormal records = 1850, Normal records = 5151, Detection time = 7.89 s, Time per record = 0.00112774443626 s Abnormal records = 2105, Normal records = 5896, Detection time = 9.02 s, Time per record = 0.00112706288695 s Abnormal records = 2357, Normal records = 6644, Detection time = 10.14 s, Time per record = 0.00112684233983 s Anomalies were detected (count = 22) [abnormal records = 2521, normal records = 7177, detection time = 10.92 s, time per record = 0.00112578553695 s] ---------------------------------------------------------------------- Applying detector to the full activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was included]... Records count: 11850 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 253, Normal records = 748, Detection time = 1.13 s, Time per record = 0.00112664413452 s Abnormal records = 479, Normal records = 1522, Detection time = 2.26 s, Time per record = 0.00113046598434 s Abnormal records = 722, Normal records = 2279, Detection time = 3.38 s, Time per record = 0.00112577366829 s Abnormal records = 971, Normal records = 3030, Detection time = 4.51 s, Time per record = 0.00112768054008 s Abnormal records = 1225, Normal records = 3776, Detection time = 5.64 s, Time per record = 0.00112878522873 s Abnormal records = 1439, Normal records = 4562, Detection time = 6.76 s, Time per record = 0.00112742733955 s Abnormal records = 1707, Normal records = 5294, Detection time = 7.89 s, Time per record = 0.00112738772801 s Abnormal records = 1956, Normal records = 6045, Detection time = 9.01 s, Time per record = 0.00112660801411 s Abnormal records = 2205, Normal records = 6796, Detection time = 10.14 s, Time per record = 0.00112647589048 s Abnormal records = 2441, Normal records = 7560, Detection time = 11.26 s, Time per record = 0.0011262802124 s Abnormal records = 2685, Normal records = 8316, Detection time = 12.39 s, Time per record = 0.00112592681971 s Anomalies were detected (count = 23) [abnormal records = 2878, normal records = 8972, detection time = 13.34 s, time per record = 0.00112551166035 s] Full working time = 6245.14 :
l_time— .te_l_time— , .te_t_time— .g_l_perc— , .g_t_perc— .f_l_perc— , .f_t_perc— .
| Type of | l_time | te_l_time | te_t_time | g_l_perc | g_t_perc | f_l_perc | f_t_perc |
|---|---|---|---|---|---|---|---|
| GNG host | 1869 | 0.39 | 4.61 | 69.5 | 78.4 | 0 | 36.9 |
| GNG host | 1839 | 0.42 | 4.72 | 60.2 | 72.5 | 0 | 47.7 |
| IGNG host | 551 | 1.02 | 11.97 | 22.2 | 26.8 | 0 | 17.3 |
| IGNG host | 567 | 1.14 | 13.34 | 28.9 | 27.0 | 0 | 20.8 |
:
- .
- .
- IGNG . , , , (, ).
- , GNG , .. 1.5-2 .
Conclusion
, . , . .
- Fast GNG .
- " " [21] [18].
- SOINN , GNG, .
- , .
- .
,
- .
- honeypot. honeypot .
- . , . , TPOT . , , .
, .
[1] http://www.irjcs.com/volumes/vol4/iss09/08.SISPCS10095.pdf "J.Rubina Parveen — Neural networks in cyber security — 2017"
[2] http://e-notabene.ru/nb/article_18834.html " .. — — 2016"
[3] https://www.sciencedirect.com/science/article/pii/S2352864816300281 "Haibo Zhang, Qing Huang, Fangwei Li, Jiang Zhu — A network security situation prediction model based on wavelet neural network with optimized parameters — 2016"
[4] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4896428/pdf/pone.0155781.pdf "Min-Joo Kang, Je-Won Kang — Intrusion Detection System Using Deep Neural Network for In-Vehicle Network Security — 2016"
[5] https://journals.nstu.ru/vestnik/download_article?id=3366 ".. , .. — — 2014"
[6] https://storage.tusur.ru/files/425/-1005__.___..pdf " .., .. — — 2013"
[7] https://www.sciencedirect.com/science/article/pii/S1877705814003579 "Halenar Igor, Juhasova Bohuslava, Juhas Martin, Nesticky Martin — Application of Neural Networks in Computer Security — 2013"
[8] http://psta.psiras.ru/read/psta2011_3_3-15.pdf " .., .., .., .. — — 2011"
[9] https://pdfs.semanticscholar.org/94f8/e1914ca526f53e9932890a0356394f9806f8.pdf "E.Kesavulu Reddy — Neural Networks for Intrusion Detection and Its Applications — 2013"
[10] http://elib.bsu.by/bitstream/123456789/179040/1/119-122.pdf " .., .., .., .. — — 2004"
[11] https://www.sans.org/reading-room/whitepapers/detection/application-neural-networks-intrusion-detection-336 "SANS Institute — Application of Neural Networks to Intrusion Detection — 2001"
[12] http://dom8a.ru/cupnewyear2014/prezentation/evlanenkova_paper.pdf " . — — ?"
[13] https://www.science-education.ru/pdf/2014/6/1632.pdf " .., .., .. — — ?"
[14] https://www.researchgate.net/publication/309038723_A_review_of_KDD99_dataset_usage_in_intrusion_detection_and_machine_learning_between_2010_and_2015 "Atilla Özgür — A review of KDD99 dataset usage in intrusion detection and machine learning between 2010 and 2015 — 2016"
[15] http://citforum.ru/security/internet/ids_overview/ " .., .. — : — 2009"
[16] https://moluch.ru/conf/tech/archive/5/1115/ " .. — . . — 2011"
[17] https://papers.nips.cc/paper/893-a-growing-neural-gas-network-learns-topologies.pdf "Fritzke B. — A Growing Neural Gas Network Learns
Topologies — 1993"
[18] https://www.mql5.com/ru/articles/163 " . — , MQL5 — 2010"
[19] https://www.researchgate.net/publication/4202425_An_incremental_growing_neural_gas_learns_topologies "Prudent Y., Ennaji A. — An incremental growing neural gas learns topologies — 2005"
[20] https://cyberleninka.ru/article/v/modifitsirovannyy-algoritm-rastuschego-neyronnogo-gaza-primenitelno-k-zadache-klassifikatsii " .., .. — , — 2014
[21] http://www.booru.net/download/MasterThesisProj.pdf "Jim Holmström — Growing Neural Gas
Experiments with GNG, GNG with Utility and Supervised GNG — 2002"
')
Source: https://habr.com/ru/post/358200/
All Articles