The solution to the problem of detecting the center line of the vessel
The essence of the task
In the process of medical diagnosis, it may be necessary to examine the vessels of the patient. Such a study is called angiography. With the advent of tomographs, in addition to classical angiography, MRI and CT angiography methods have appeared, which, in contrast to traditional angiography, which gives only a flat image in one projection, allows us to obtain a complete three-dimensional representation of the vessels. To carry out such studies, a contrast is injected into the patient’s blood — a special substance that makes the vessels in the pictures brighter. Depending on the intended diagnosis, the doctor either assesses the overall picture, or tries to find specific areas of the vessels in which problems have arisen. If a portion of the vessel is narrowed and transmits less blood than it should, then this place is called stenosis.
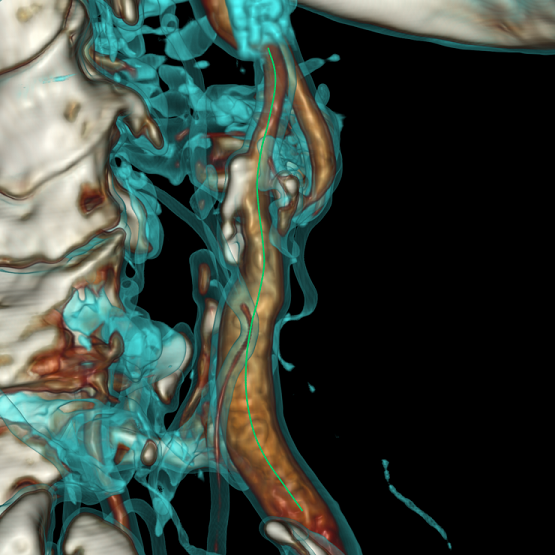
One of the tasks of a doctor is to find stenoses and assess how dangerous they are. The task of the developer, as usual, to facilitate the work of the end user. To do this, it is necessary to build a full 3D model of the vessel walls and conduct their primary analysis. This is a big and interesting task, however, it is based on a more simple and well-known problem - the construction of the central line of the vessel.
First try
Before continuing to read this article, it is advisable to get a little familiar with the presentation of the data obtained from the work of tomographs. This can be done by reading our long-written article about the voxel render of the DICOM Viewer from the inside . In short: there is a 3D array of numbers, in each element of which the signal value (intensity) is stored. This array is called volume. The element itself is a voxel, and its indices in the 3D array will be 3D coordinates. Before the rendering of each voxel, its intensity is processed by the function, and the voxel acquires a certain color, brightness and transparency.
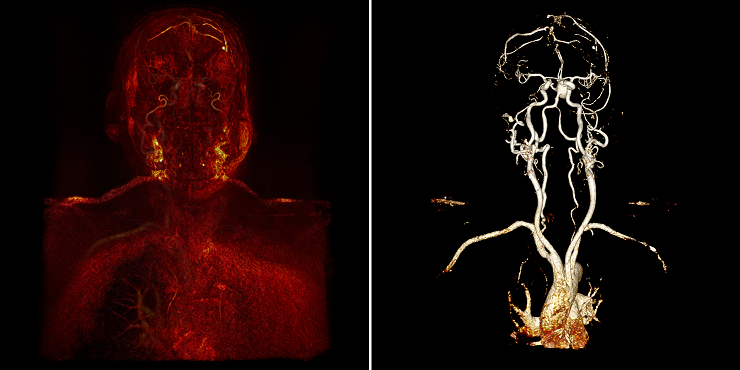
Regarding the task, the first thing I encountered was that our render allows us to show all the vessels at the visual level. Namely, from an incomprehensible “porridge”, as in the figure on the left, playing around with the settings you can make quite obvious vessels, as in the figure on the right:
')
It seems that the problem is solved: “here they are” vessels, a region growing algorithm immediately comes to mind: we know the color and transparency of the voxels we need, which means we can iterate through their neighbors until the path between the specified points has been laid.
Segmentation vessel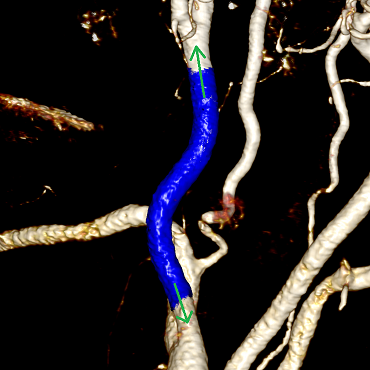

In addition, the idea comes that if you deal with the branches, you can extend the entire network of vessels. But the reality is not so simple. The vessel can fit closely to the bones (their voxels are identical in color, so the bone will be perceived as part of the vessel):
Example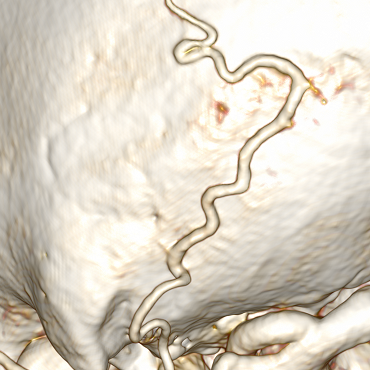

The vessel may curl or fit close to itself:
Example

Parts of the vessel can be very thin and even interrupted:
Example

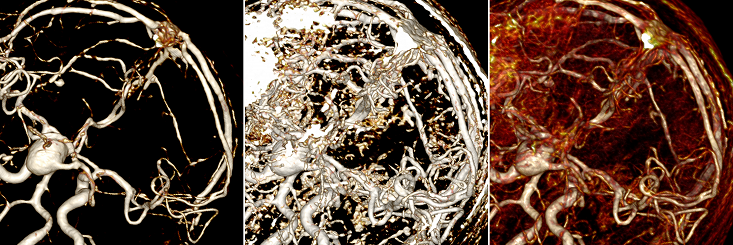
Finding optimal render settings is nontrivial. The user can change the so-called window, and there is no specific value of the window suitable for all vessels of at least the same study. The window has to be customized each time for a specific case. Below are three options for the same place with different settings:
As a result, we come to the fact that the intuitively seemingly correct algorithm needs to either be refined, or a completely different approach should be tried. After a lot of unsuccessful attempts, I had to try something else.
Classic approach
The main approach to the problem of vessel detection (vessel detection) is to calculate the eigenvalues of the Hessian matrix. Also recently, they are trying to connect neural networks to this task. However, the standard solution looks more reliable because it is mentioned in more literature and is used not only for finding vessels, but also for many other tasks (for example, detecting objects in astronomical images), so neural networks had to be abandoned.
In our case, the Hesse matrix is a matrix whose elements are partial derivatives of the second order of intensity in a particular voxel:
H_ {M} = \ begin {pmatrix} \ frac {\ partial ^ 2 f} {\ partial ^ 2 x} & \ frac {\ partial ^ 2 f} {\ partial x \ partial y} & \ frac {\ partial ^ 2 f} {\ partial x \ partial z} \\ \ frac {\ partial ^ 2 f} {\ partial x \ partial y} & \ frac {\ partial ^ 2 f} {\ partial ^ 2 y} & \ frac {\ partial ^ 2 f} {\ partial y \ partial z} \\ \ frac {\ partial ^ 2 f} {\ partial x \ partial z} & \ frac {\ partial ^ 2 f} {\ partial y \ partial z} & \ frac {\ partial ^ 2 f} {\ partial ^ 2 z} \ end {pmatrix}
After constructing the Hesse matrix, it is necessary to find its eigenvalues and eigenvectors by solving the following equation:
\ begin {vmatrix} \ frac {\ partial ^ 2 f} {\ partial ^ 2 x} - \ lambda_ {1} & \ frac {\ partial ^ 2 f} {\ partial x \ partial y} & \ frac { \ partial ^ 2 f} {\ partial x \ partial z} \\ \ frac {\ partial ^ 2 f} {\ partial x \ partial y} & \ frac {\ partial ^ 2 f} {\ partial ^ 2 y} - \ lambda_ {2} & \ frac {\ partial ^ 2 f} {\ partial y \ partial z} \\ \ frac {\ partial ^ 2 f} {\ partial x \ partial z} & \ frac {\ partial ^ 2 f} {\ partial y \ partial z} & \ frac {\ partial ^ 2 f} {\ partial ^ 2 z} - \ lambda_ {3} \ end {vmatrix} = 0
p1 = A(1,2)^2 + A(1,3)^2 + A(2,3)^2 if (p1 == 0) eig1 = A(1,1) eig2 = A(2,2) eig3 = A(3,3) else q = trace(A)/3 p2 = (A(1,1) - q)^2 + (A(2,2) - q)^2 + (A(3,3) - q)^2 + 2 * p1 p = sqrt(p2 / 6) B = (1 / p) * (A - q * I) r = det(B) / 2 if (r <= -1) phi = pi / 3 else if (r >= 1) phi = 0 else phi = acos(r) / 3 end eig1 = q + 2 * p * cos(phi) eig3 = q + 2 * p * cos(phi + (2*pi/3)) eig2 = 3 * q - eig1 - eig3 end Where A is the original 3x3 matrix; I - 3x3 identity matrix; eig1, eig2, eig2 - eigenvalues; trace () - returns the trace of the matrix; det () - returns the determinant of the matrix.
Now we can find eigenvectors. To do this in the equation:
\ begin {vmatrix} \ frac {\ partial ^ 2 f} {\ partial ^ 2 x} - \ lambda & \ frac {\ partial ^ 2 f} {\ partial x \ partial y} & \ frac {\ partial ^ 2 f} {\ partial x \ partial z} \\ \ frac {\ partial ^ 2 f} {\ partial x \ partial y} & \ frac {\ partial ^ 2 f} {\ partial ^ 2 y} - \ lambda & \ frac {\ partial ^ 2 f} {\ partial y \ partial z} \\ \ frac {\ partial ^ 2 f} {\ partial x \ partial z} & \ frac {\ partial ^ 2 f} {\ partial y \ partial z} & \ frac {\ partial ^ 2 f} {\ partial ^ 2 z} - \ lambda \ end {vmatrix} \ cdot \ begin {vmatrix} V_ {x} \\ V_ {y} \\ V_ {z} \ end {vmatrix} = 0
substitute one of the found eigenvalues instead of 
If the voxel belongs to a vessel, the so-called vascularity is calculated on the basis of the eigenvalues of the matrix. There are a lot of different formulas in the literature, and we adapted one of them for ourselves:
The more vascular, the closer the voxel to the center of the vessel. Now it is easy to imagine a simple algorithm for finding the center line from a given voxel:
- Determine the direction of the vessel in the current voxel,
- Make a cut of the vessel perpendicular to the direction and move along the gradient to the cut voxel with maximum vascularity,
- Determine the direction of the vessel in the voxel with maximum vascularity,
- We take a small step in the direction of the vessel and get into the new voxel,
- Go back to step 1.
From the original point, the movement occurs in two directions, because we cannot know in advance which of them we need:
Algorithm animation

As a result, we get not the most beautiful, but the correct center line:
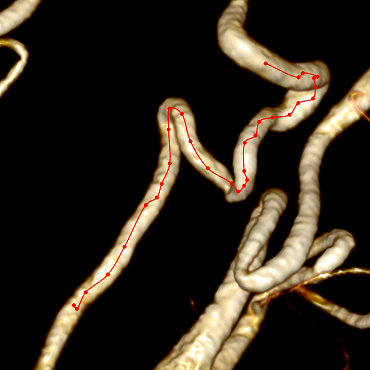
To make the center line less angular, you need to abandon the integer coordinates of the voxels and move to the fractional coordinates of points in 3D space; you can also perform a slight smoothing of the center line after construction.
To go to the fractional coordinates, we used the bicubic interpolation when obtaining the intensity values. The general equation of the filter in one-dimensional space is as follows:
Where
Consider the case for 1D. If given values are in points
Total predicted value:
Now consider our full case for 3D space. As you might guess, we will interpolate inside the cube with the dimension of 4x4x4 voxel. We take as a basis the weights considered for the 1D case:
Where
It may seem interesting to someone that the sum of the weights of all voxels is 1:
Details
Let us return to the calculation of derivatives for the Hesse matrix. We have coordinates and intensity, as the second partial derivative is numerically considered as everyone knows: the basic is the finite difference method. For point
Formula
To eliminate noise, before calculating the derivatives, it is necessary to perform a Gaussian smoothing with a certain value.
Where
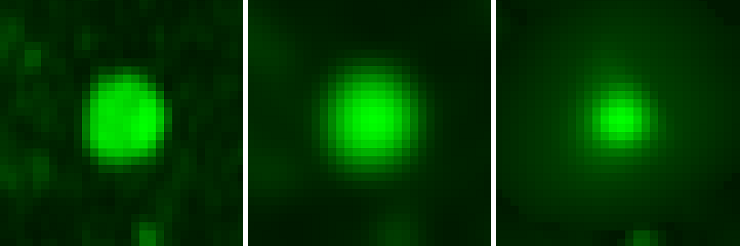
The intensity values in the perpendicular section of the vessel before smoothing, after smoothing, in the third figure, the values of vascularity:
In this case, the greater the value
However, with increasing
We used the so-called gamma normalization:
Now, if we try to calculate the value of vascularity with a small
The calculation algorithm for an arbitrary point when building the center line looks like this:
- select value
and perform smoothing at a point and in its neighbors (they are needed to calculate the derivatives),
- calculate the second derivatives and build the Hesse matrix,
- normalize the Hesse matrix by multiplying it by
, and find eigenvalues,
- calculate vascularity
- if there are unused
then go back to the beginning, otherwise go to point 2.
- select value
- Calculate the eigenvectors of the Hesse matrix, for which the vascularity turned out to be maximum, and obtain the vector-direction of the vessel.
Combining this algorithm with the algorithm for building the center line, we get the final result:
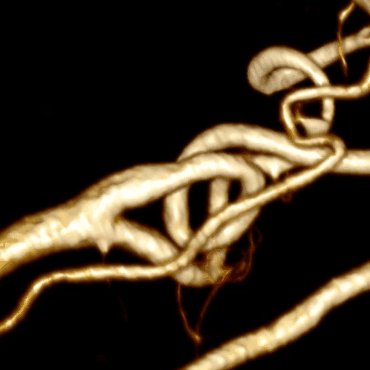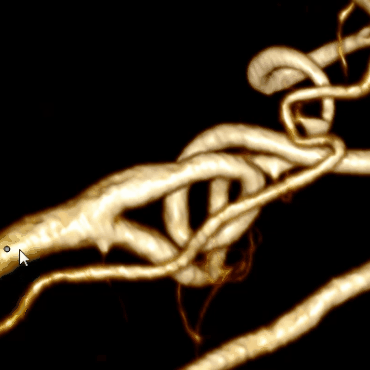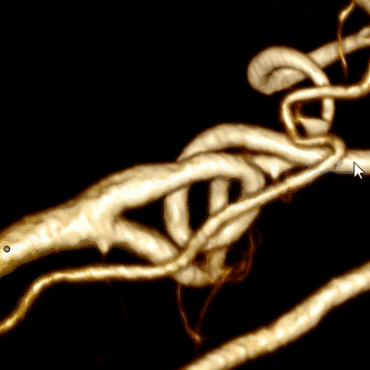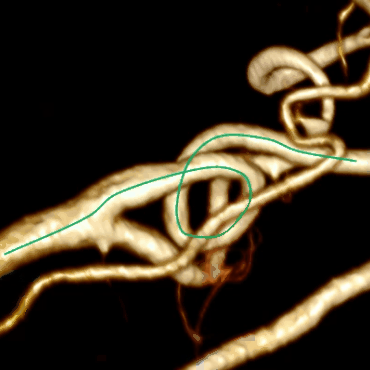
Optimization
The approach described above will work fine, but there are a few points. First, to improve performance, smoothing must be performed in advance for all voxels and for all
The idea is that since after each smoothing, the intensity values in the neighboring voxels blur and become approximately equal, there is no point in storing them all. Those. the more
In general, it works like this. The zero volume layer is original. A smoothing filter (¼, ½, ¼) is applied several times along each of the volume axes, and after that the dimensionality of the volume is halved. The resulting volume will belong to the first layer. Then the operation is repeated and the second layer is obtained. Then the third, etc. Each layer corresponds to a specific value.
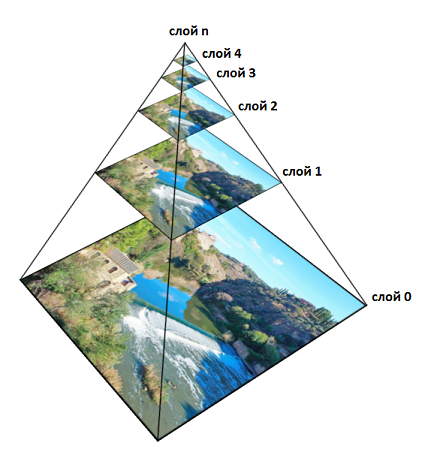
It is easy to calculate that in 3D the total amount of memory will be equal to
Total
It can be considered that the described approach to building the center line is extremely effective, but it has some drawbacks:
- For CT studies, it is not possible to detect vessels that pass inside the bones (as, for example, in the spine)
- It is not always possible to detect vessels if they pass near elongated elongated bones, tissues or structures (in such cases bones, tissues and structures themselves may be mistaken for vessels)
- The bottleneck is bifurcations (bifurcation of vessels)
Problems 1 and 2 are solved by subtracting the bones. For this, two CT examinations are necessary - with and without contrast. It is clear that the only difference between such studies will be illuminated vessels. In other words, if we subtract the intensity of each voxel of a study without vessels from the intensity of each voxel of a study with vessels, then as a result only vessels related to vessels will have non-zero intensity. But since the two studies cannot be done with a completely identical position of the patient’s body, the main problem is the combination of the two studies in 3D space. To do this, use the transformation of rotation and displacement. Orientation in space goes on the bones, because they are rigid structures. To find the bones, we used a very interesting algorithm of waterfalls based on graphs (waterfall based on graphs).
The problem of bifurcations is solved by specifying the starting and ending points of the vessel, between which it is necessary to build a central line. It needs to start building at each of the two points in each of the two directions, and when crossing the central lines, simply merge them into one. Since vessels branching in only one direction (thicker ones are broken up into smaller ones in the case of arteries, and thinner ones combine to thicker ones in the case of veins), then this approach allows connecting two given points.
That's all, thank you for your attention. Just in case a link to the product .
Source: https://habr.com/ru/post/354702/
All Articles