ggplot2: how to easily combine multiple graphs in one, part 3
This article will show step by step how to combine several ggplot graphs in one or several illustrations using the helper functions available in the R ggpubr , cowplot and gridExtra packages . We also describe how to export the resulting graphs to a file.
First part
The second part of
In this section, we will show how to display a table and text along with a graph. We will use the iris dataset.
Start by creating these graphs:
We conclude by combining all three graphs using the
')

To add tables, graphs or other elements based on tables to the ggplot workspace, there is a function
We use the density.p and stable.p graphics created in the previous section.

Since the scatterplot placed inside is superimposed at several points, a transparent background is used for it.

Import background image. Use either the
To test the example below, make sure the png package is installed. You can install it using the
Combine ggplot with background image. R function:
Change the transparency of the fill in the scatter diagrams by specifying the argument alpha. The value must be between [0, 1], where 0 is full transparency, and 1 is lack of transparency.
Another example, overlaying a map of France on ggplot2:

If you have a long list of ggplot-s, say, n = 20, you might want to organize them by placing them on several pages. If there are 4 graphics on the page, for 20 you will need 5 pages.
The
For example, the code below
returns two pages with two graphs on each. You can display each page like this:
Ordered graphs can be exported to a pdf file using the
PDF file
Multipage output can be obtained with the
Let us arrange the graphs created in the previous sections.
Function R:
First, create a list of 4 ggplot-s corresponding to the variables Sepal.Length, Sepal.Width, Petal.Length and Petal.Width in the iris data set.
You can then export individual graphs to a file (pdf, eps or png) (one graph per page). You can arrange the graphics (2 per page) when exporting.
Export individual graphs to pdf (one per page):
Arrange and export. Set nrow and ncol to display several graphs on the same page:
First part
The second part of
Mix table, text and ggplot2 graphics
In this section, we will show how to display a table and text along with a graph. We will use the iris dataset.
Start by creating these graphs:
- density graph of Sepal.Length. R function:
ggdensity()[in ggpubr ] - summary table containing descriptive statistics (mean, standard deviation, etc.) Sepal.Length
- Function R for calculating descriptive statistics:
desc_statby()[in ggpubr ] - R function to create a table with text:
ggtexttable()[in ggpubr ]
- Function R for calculating descriptive statistics:
- paragraph text . R function:
ggparagraph()[in ggpubr ]
We conclude by combining all three graphs using the
ggarrange() function [in ggpubr ].')
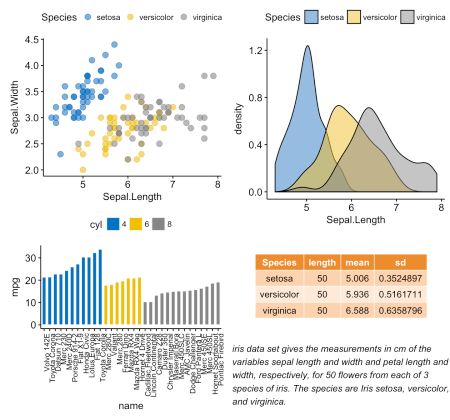
# "Sepal.Length" #:::::::::::::::::::::::::::::::::::::: density.p <- ggdensity(iris, x = "Sepal.Length", fill = "Species", palette = "jco") # Sepal.Length #:::::::::::::::::::::::::::::::::::::: # stable <- desc_statby(iris, measure.var = "Sepal.Length", grps = "Species") stable <- stable[, c("Species", "length", "mean", "sd")] # , "medium orange" ( ) stable.p <- ggtexttable(stable, rows = NULL, theme = ttheme("mOrange")) # #:::::::::::::::::::::::::::::::::::::: text <- paste("iris data set gives the measurements in cm", "of the variables sepal length and width", "and petal length and width, respectively,", "for 50 flowers from each of 3 species of iris.", "The species are Iris setosa, versicolor, and virginica.", sep = " ") text.p <- ggparagraph(text = text, face = "italic", size = 11, color = "black") # ggarrange(density.p, stable.p, text.p, ncol = 1, nrow = 3, heights = c(1, 0.5, 0.3)) 
Adding a graphic element to ggplot
To add tables, graphs or other elements based on tables to the ggplot workspace, there is a function
annotation_custom() [in ggplot2 ]. Simplified format: annotation_custom(grob, xmin, xmax, ymin, ymax) - grob : external graphic element to display
- xmin , xmax : x-location in coordinates (horizontal location)
- ymin , ymax : y-location in coordinates (vertical location)
Place the table in ggplot
We use the density.p and stable.p graphics created in the previous section.
density.p + annotation_custom(ggplotGrob(stable.p), xmin = 5.5, ymin = 0.7, xmax = 8) 
Place the scatterplot in ggplot
- Create a scatter plot for y = “Sepal.Width” by x = “Sepal.Length” from the iris dataset. R:
ggscatter()[ ggpubr ] function . - Create a separate scatterplot of x and y variables with a transparent background. R function:
ggboxplot()[ ggpubr ]. - Transform a scatterplot into a graphical object called “grob” in Grid terminology. R function:
ggplotGrob()[ ggplot2 ]. - Place the scatter plots inside the scatter plots. R:
annotation_custom()[ ggplot2 ] function .
Since the scatterplot placed inside is superimposed at several points, a transparent background is used for it.
# ("Species") #:::::::::::::::::::::::::::::::::::::::::::::::::::::::::: sp <- ggscatter(iris, x = "Sepal.Length", y = "Sepal.Width", color = "Species", palette = "jco", size = 3, alpha = 0.6) # x/y #:::::::::::::::::::::::::::::::::::::::::::::::::::::::::: # x xbp <- ggboxplot(iris$Sepal.Length, width = 0.3, fill = "lightgray") + rotate() + theme_transparent() # ybp <- ggboxplot(iris$Sepal.Width, width = 0.3, fill = "lightgray") + theme_transparent() # # “grob” Grid xbp_grob <- ggplotGrob(xbp) ybp_grob <- ggplotGrob(ybp) # #:::::::::::::::::::::::::::::::::::::::::::::::::::::::::: xmin <- min(iris$Sepal.Length); xmax <- max(iris$Sepal.Length) ymin <- min(iris$Sepal.Width); ymax <- max(iris$Sepal.Width) yoffset <- (1/15)*ymax; xoffset <- (1/15)*xmax # xbp_grob sp + annotation_custom(grob = xbp_grob, xmin = xmin, xmax = xmax, ymin = ymin-yoffset, ymax = ymin+yoffset) + # ybp_grob annotation_custom(grob = ybp_grob, xmin = xmin-xoffset, xmax = xmin+xoffset, ymin = ymin, ymax = ymax) 
Add a background image to ggplot2 graphics
Import background image. Use either the
readJPEG() function [in the jpeg package] or the readPNG() function [in the png package] depending on the format of the background image.To test the example below, make sure the png package is installed. You can install it using the
install.packages(“png”) command. # img.file <- system.file(file.path("images", "background-image.png"), package = "ggpubr") img <- png::readPNG(img.file) Combine ggplot with background image. R function:
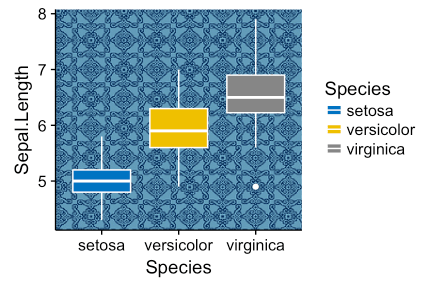
background_image() [in ggpubr ]. library(ggplot2) library(ggpubr) ggplot(iris, aes(Species, Sepal.Length))+ background_image(img)+ geom_boxplot(aes(fill = Species), color = "white")+ fill_palette("jco") 
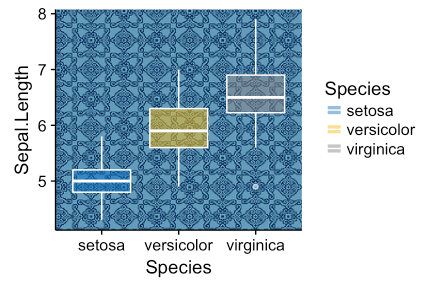
Change the transparency of the fill in the scatter diagrams by specifying the argument alpha. The value must be between [0, 1], where 0 is full transparency, and 1 is lack of transparency.
library(ggplot2) library(ggpubr) ggplot(iris, aes(Species, Sepal.Length))+ background_image(img)+ geom_boxplot(aes(fill = Species), color = "white", alpha = 0.5)+ fill_palette("jco") 
Another example, overlaying a map of France on ggplot2:
mypngfile <- download.file("https://upload.wikimedia.org/wikipedia/commons/thumb/e/e4/France_Flag_Map.svg/612px-France_Flag_Map.svg.png", destfile = "france.png", mode = 'wb') img <- png::readPNG('france.png') ggplot(iris, aes(x = Sepal.Length, y = Sepal.Width)) + background_image(img)+ geom_point(aes(color = Species), alpha = 0.6, size = 5)+ color_palette("jco")+ theme(legend.position = "top") 
We have graphics on several pages.
If you have a long list of ggplot-s, say, n = 20, you might want to organize them by placing them on several pages. If there are 4 graphics on the page, for 20 you will need 5 pages.
The
ggarrange() function [in ggpubr ] provides a convenient solution to place several ggplot-s on several pages. After setting the nrow and ncol arguments, the ggarrange() function automatically calculates the number of pages it takes to place all the graphs. It returns a list of ordered ggplot-s.For example, the code below
multi.page <- ggarrange(bxp, dp, bp, sp, nrow = 1, ncol = 2) returns two pages with two graphs on each. You can display each page like this:
multi.page[[1]] # 1 multi.page[[2]] # 2 Ordered graphs can be exported to a pdf file using the
ggexport() function [in ggpubr ]: ggexport(multi.page, filename = "multi.page.ggplot2.pdf") PDF file
Multipage output can be obtained with the
marrangeGrob() function [in gridExtra ]. library(gridExtra) res <- marrangeGrob(list(bxp, dp, bp, sp), nrow = 1, ncol = 2) # pdf- ggexport(res, filename = "multi.page.ggplot2.pdf") # res Nested positioning with ggarrange ()
Let us arrange the graphs created in the previous sections.
p1 <- ggarrange(sp, bp + font("x.text", size = 9), ncol = 1, nrow = 2) p2 <- ggarrange(density.p, stable.p, text.p, ncol = 1, nrow = 3, heights = c(1, 0.5, 0.3)) ggarrange(p1, p2, ncol = 2, nrow = 1) 
Export graphs
Function R:
ggexport() [in ggpubr ].First, create a list of 4 ggplot-s corresponding to the variables Sepal.Length, Sepal.Width, Petal.Length and Petal.Width in the iris data set.
plots <- ggboxplot(iris, x = "Species", y = c("Sepal.Length", "Sepal.Width", "Petal.Length", "Petal.Width"), color = "Species", palette = "jco" ) plots[[1]] # plots[[2]] # .. You can then export individual graphs to a file (pdf, eps or png) (one graph per page). You can arrange the graphics (2 per page) when exporting.
Export individual graphs to pdf (one per page):
ggexport(plotlist = plots, filename = "test.pdf") Arrange and export. Set nrow and ncol to display several graphs on the same page:
ggexport(plotlist = plots, filename = "test.pdf", nrow = 2, ncol = 1) Source: https://habr.com/ru/post/339090/
All Articles