Progress Tracking in R
It doesn't matter if we give ourselves an account of this, but when we have to wait, we worry and burn with impatience. This is especially true of waiting "blindly", that is, when it is unknown how much more will have to suffer. As Brad Allan Myers, who is considered the inventor of the state indicator in the 1980s, found out, the ability to track progress while waiting can significantly improve the user interaction mechanism with an application (Myers, 1985).

Typical Status Indicator by Simeon87 [ GPL ], Wikimedia Commons
Since I program in R for research in bioinformatics, my code is usually not for the general public, but it’s still important that my users, I mean colleagues and researchers, are as happy as possible. But tracking progress in R is not an easy task. This article presents several possible solutions, including my own ( pbmcapply ).
The easiest way to track the progress in R is to periodically display the percentage of work completed at the moment: by default, on the screen, or write it to some file with logs somewhere on the disk. It makes no sense to say that this is probably the least elegant way to solve, but many still use it.
')
The best (and still simple) solution is to include the pbapply package. If you believe the information on his page , the package is very popular - 90 thousand downloads. Its easy to use. Whenever you are going to call apply, call its pbapply version . For example:
While the numbers are being processed, a periodically updated status indicator will be displayed.

Pbapply generated status indicator. The user can see how much more time is needed, and the current state in the form of an indicator.
Although pbapply is a great tool, and I use it often, I wasn’t able to display progress for the parallel version of apply - mcapply - until recently. In September, the author pbapply added support for clusters in a simple network (snow-type — Simple Network Of Workstations — simple network of workstations) and multi-core branching to his package. But this approach involves the separation of elements into parts and the consistent application of mcapply to them. One of the drawbacks of this approach is that if the number of elements greatly exceeds the number of cores, you need to call mcapply many times. The mcapply calls built on Unix / Linux fork () are very expensive: branching into many child processes takes time and too much memory.
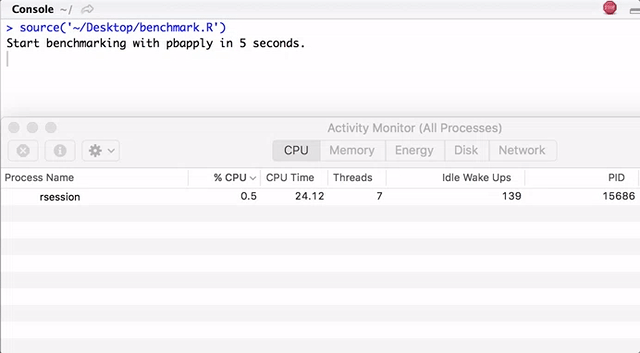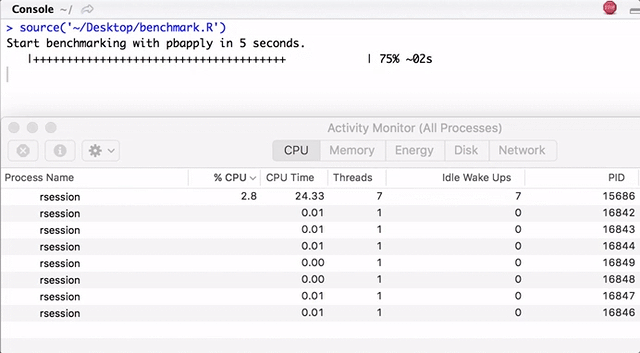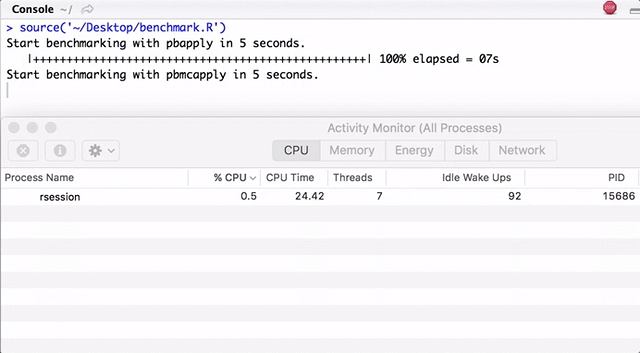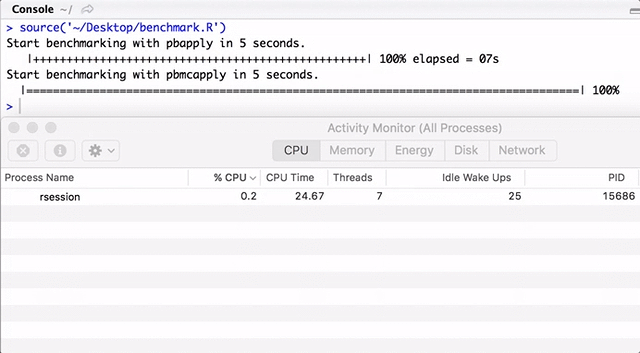
Note that pbapply generates many child processes, and pbmcapply re-uses them where possible. For pbapply / pbmcapply 4 cores are allocated. The code for R can be downloaded from here .
Pbmcapply - my own solution to the problem. It is available through the CRAN package, and is easy to use:
As the name implies, I was inspired by the pbapply package. Unlike pbapply , my solution does not involve many calls to mcapply . Pbmcapply uses the future package instead.

Pbmcapply schema. FutureCall () is executed in a separate process, which then forks into a specified number of children. Child processes periodically transmit information about their progress through progressMonitor. As soon as progressMonitor receives data, it displays the status on the standard output device.
In computer science, future denotes an object that will later contain a value. This allows the program to execute some code as future and, without waiting for the output, go to the next step. In pbmcapply, mcapply can be placed in the future. Future will periodically provide information about its state to the main program, which, in turn, will display a status indicator.
Since the pbmcapply resource costs are minimal and nonlinear, we get a significant performance improvement when the number of elements greatly exceeds the number of processor cores. As an illustration, single- and multi-threaded apply-functions are used in R. Obviously, even with pbmcapply , performance suffers, since it takes time to start the monitoring process.

Performance comparison pbapply and pbmcapply. The code for R can be downloaded from here . The left panel shows the cost of resources when calling each of the packages. The right panel shows the time for each call.
Everything has its price. While enjoying the convenience of interactive state tracking, remember this slows down the program a bit.
As always, there is no universal solution. If your main priority is performance (for example, when you run a program in a cluster), perhaps the best way to track progress is print . However, if the extra few seconds do not solve anything, please use my solution ( pbmcapply ) or pbapply to make your program more convenient.
Myers, BA (1985). For computer-human interfaces. In ACM SIGCHI Bulletin (Vol. 16, No. 4, pp. 11–17). ACM.

Typical Status Indicator by Simeon87 [ GPL ], Wikimedia Commons
Since I program in R for research in bioinformatics, my code is usually not for the general public, but it’s still important that my users, I mean colleagues and researchers, are as happy as possible. But tracking progress in R is not an easy task. This article presents several possible solutions, including my own ( pbmcapply ).
Output on display
The easiest way to track the progress in R is to periodically display the percentage of work completed at the moment: by default, on the screen, or write it to some file with logs somewhere on the disk. It makes no sense to say that this is probably the least elegant way to solve, but many still use it.
')
Pbapply
The best (and still simple) solution is to include the pbapply package. If you believe the information on his page , the package is very popular - 90 thousand downloads. Its easy to use. Whenever you are going to call apply, call its pbapply version . For example:
# , nums <- 1:10 # sapply, sqrt <- sapply(nums, sqrt) # pbapply sqrt <- pbsapply(nums, sqrt) While the numbers are being processed, a periodically updated status indicator will be displayed.

Pbapply generated status indicator. The user can see how much more time is needed, and the current state in the form of an indicator.
Although pbapply is a great tool, and I use it often, I wasn’t able to display progress for the parallel version of apply - mcapply - until recently. In September, the author pbapply added support for clusters in a simple network (snow-type — Simple Network Of Workstations — simple network of workstations) and multi-core branching to his package. But this approach involves the separation of elements into parts and the consistent application of mcapply to them. One of the drawbacks of this approach is that if the number of elements greatly exceeds the number of cores, you need to call mcapply many times. The mcapply calls built on Unix / Linux fork () are very expensive: branching into many child processes takes time and too much memory.

Note that pbapply generates many child processes, and pbmcapply re-uses them where possible. For pbapply / pbmcapply 4 cores are allocated. The code for R can be downloaded from here .
Pbmcapply
Pbmcapply - my own solution to the problem. It is available through the CRAN package, and is easy to use:
# pbmcapply install.packages("pbmcapply") As the name implies, I was inspired by the pbapply package. Unlike pbapply , my solution does not involve many calls to mcapply . Pbmcapply uses the future package instead.

Pbmcapply schema. FutureCall () is executed in a separate process, which then forks into a specified number of children. Child processes periodically transmit information about their progress through progressMonitor. As soon as progressMonitor receives data, it displays the status on the standard output device.
In computer science, future denotes an object that will later contain a value. This allows the program to execute some code as future and, without waiting for the output, go to the next step. In pbmcapply, mcapply can be placed in the future. Future will periodically provide information about its state to the main program, which, in turn, will display a status indicator.
Since the pbmcapply resource costs are minimal and nonlinear, we get a significant performance improvement when the number of elements greatly exceeds the number of processor cores. As an illustration, single- and multi-threaded apply-functions are used in R. Obviously, even with pbmcapply , performance suffers, since it takes time to start the monitoring process.

Performance comparison pbapply and pbmcapply. The code for R can be downloaded from here . The left panel shows the cost of resources when calling each of the packages. The right panel shows the time for each call.
Everything has its price. While enjoying the convenience of interactive state tracking, remember this slows down the program a bit.
Conclusion
As always, there is no universal solution. If your main priority is performance (for example, when you run a program in a cluster), perhaps the best way to track progress is print . However, if the extra few seconds do not solve anything, please use my solution ( pbmcapply ) or pbapply to make your program more convenient.
Links
Myers, BA (1985). For computer-human interfaces. In ACM SIGCHI Bulletin (Vol. 16, No. 4, pp. 11–17). ACM.
Source: https://habr.com/ru/post/317314/
All Articles