"Pledged by nature": DNA-based data storage system

/ photo by MIKI Yoshihito CC
In our blog on Habré, we regularly share with you the latest news from the world of IaaS. For example, we recently talked about what changes are waiting for data centers in the future. We also talked about how large Internet companies store their data. Today we would again like to turn to the topic of data storage and tell you about a promising development - DNA-based repositories.
Hard drives, which are widely used in data centers in the world for data storage, are not famous for their durability. The Backblaze team conducted a study and found out that HDDs store information for only 10 years.
')
Unfortunately, these are modern realities - storage devices cannot last forever. For this reason, researchers from around the world are trying to find a way to store data for as long as possible — ideally, infinitely.
And they found him. It is believed that the answer to all questions lies in the DNA - it has a high recording density (1 exabyte per 1 mm3) and durability (the established decay period is more than 500 years).
The size of the “digital universe” will exceed 16 zettabytes by 2017. A large proportion of this data is stored as archives. For example, the company Facebook has recently built a separate data center for the "cold" storage of 1 exabyte of data. The same amount of information can fit in 1 mm3 of DNA.
Data storage in DNA takes place in three stages: the conversion of digital data into a DNA nucleotide sequence, the synthesis of DNA molecules and, directly, the storage of data. In order for the data to be counted, it is necessary to isolate the required sequence from the DNA molecule and convert it to its original form.
It is worth noting that there are some difficulties in working with DNA repositories, for example, there are questions about the cost of data encryption, but researchers are sure that as medical technology develops, it will decline.
This is what happens. The time to carry out the synthesis and sequencing decreases exponentially, and the growth of their efficiency follows Moore's law.
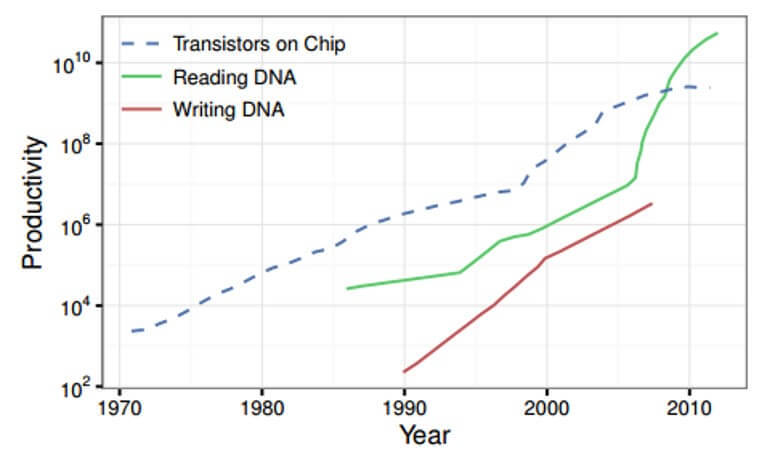
Trends in DNA synthesis in comparison with the increase in the number of transistors on a chip
Moreover, the cost of such a procedure also falls. Previously, the cost of deciphering the human genome was millions of dollars, and today it has dropped to a few hundred.
These positive trends have led scientists from the University of Washington to begin developing a DNA-based key-value data storage system. They want to explore the possibility of using such systems in modern architectures.
Information coding process
DNA contains four types of nucleotides: adenine (A), cytosine (C), guanine (G) and thymine (T). DNA strand is a linear sequence of these nucleotides. Thus, we have four code symbols (A, C, G, and T), so the obvious approach to storing binary data is to encode them in quaternary notation, for example, 0 = A, 1 = C, 2 = G, and 3 = T. However, it should be borne in mind that the synthesis and sequencing are error prone.
The probability of errors can be reduced if the binary information is encoded not in quaternary, but in the ternary number system, as shown in the figure below. To avoid inefficient conversion of the original binary data to the ternary number system, the Huffman code is used.
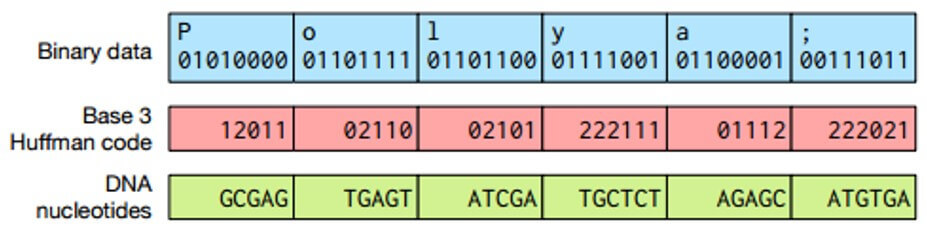
Comparison of binary data and DNA nucleotides
Each of the three digits corresponds to the DNA nucleotide in accordance with the table (below), and the nucleotides in the chain do not repeat, which leads to a decrease in sequencing error.
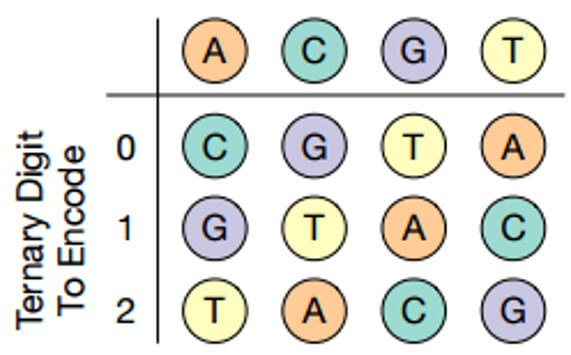
Nucleotide Coding Table
To ensure the possibility of random access to data, scientists have oganized the translation of keys into unique sequences of primers. Primers are short synthetic strands that define the beginning and end of the region to be amplified.
Primers provide random access using a polymerase chain reaction that generates multiple copies of DNA in a solution. The chains of a particular object have a common primer, and different chains with the same primer differ in address.
“By controlling the sequences that are used as primers for the polymerase chain reaction (PCR), we can indicate which strands in the solution will undergo amplification. In order to read the key value in the solution, we simply perform PCR using the primer corresponding to this key, ”say the scientists.
DNA storage system
A DNA-based storage system consists of a DNA synthesizer that encodes data, a data storage container and a DNA sequencer that reads DNA sequences and translates them back into a “number”.
The process of reading and writing data in short form is shown in the diagram below.
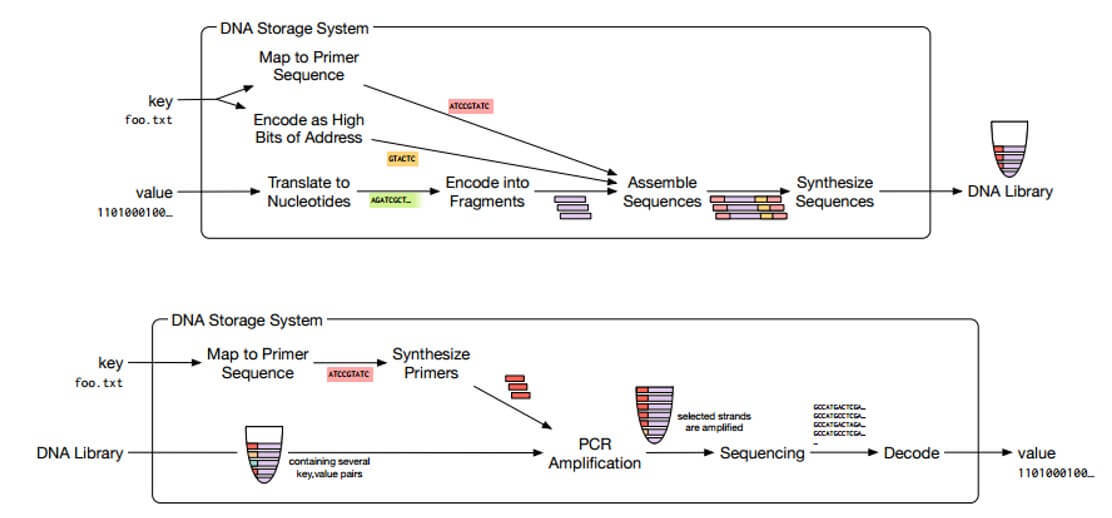
Work of DNA-based storage systems
During reading, DNA samples are removed from the pool, reducing the amount of DNA available for subsequent operations. But DNA is easily copied, so if necessary, pools can easily fill in the missing fragments. If there are difficulties in sequential amplification, DNA synthesis can be re-performed in the pool after reading.
Conclusion
In the future, such systems will potentially allow you to store a huge amount of data on microscopic media. Imagine a “flash drive” with a capacity of 100 mm3, capable of storing about 100,000 PB bytes of data.
However, so far the biggest obstacle to the introduction of such technologies remains time. Decoding and reading the DNA molecule takes many hours. Therefore, this type of storage is hardly suitable for the content of frequently used data, but it can turn our understanding of long-term storage in data centers.
PS Other materials on the topic from our blog on Habré:
- DNA and data storage solution
- Data Storage: What is the future?
- Scientists again "puzzled": Is it possible to upgrade the brain and what to expect from it
- Working with data: How do large companies
PPS We have prepared links to practical manuals in case you have time for the weekend to get acquainted with our IaaS-provider 1cloud and test its capabilities:
Source: https://habr.com/ru/post/306656/
All Articles