Strategies to speed up code on R, part 2
The for loop in R can be very slow if it is applied in its pure form, without optimization, especially when it comes to dealing with large data sets. There are a number of ways to make your code faster, and you will probably be surprised to know how much.
This article describes several approaches, including simple changes in logic, parallel processing and
Let's try to speed up the code with a for loop and a conditional operator (if-else) to create a column that is added to the data set (data frame, df). The code below creates this initial data set.
In the first part : vectorization, only true conditions, ifelse.
In this part: which, apply, byte-by-byte compilation, Rcpp, data.table, results.
')
Using the
We use the

Using apply and for loops in R
This is probably not the best example to illustrate the effectiveness of byte-based compilation, since the resulting time is slightly higher than the usual form. However, for more complex functions, byte compilation has proven to be effective. I think it is worth trying on occasion.

Apply, for loop and byte code compilation
Let's go to a new level. Before that, we increased speed and performance with various strategies and found that using
Below is the same logic implemented in C ++ using the Rcpp package. Save the code below as “MyFunc.cpp” in your working session R session directory (or you will have to use sourceCpp using the full path). Note that the comment
Parallel processing:
Remove more unnecessary objects in the code with
Dataframe and data.table
Method: Speed, number of lines in df / elapsed time = n lines per second
Source: 1X, 120000 / 140.15 = 856.2255 rows per second (normalized to 1)
Vectorized: 738X, 120000 / 0.19 = 631578.9 lines per second
Only true conditions: 1002X, 120000 / 0.14 = 857142.9 lines per second
ifelse: 1752X, 1200000 / 0.78 = 1,500,000 lines per second
which: 8806X, 2985984 / 0.396 = 7540364 lines per second
Rcpp: 13476X, 1200000 / 0.09 = 11538462 lines per second
The numbers above are approximate and based on random runs. There are no calculations of the results for
This article describes several approaches, including simple changes in logic, parallel processing and
Rcpp , increasing the speed by several orders of magnitude, so that 100 million rows of data or even more can be processed normally.Let's try to speed up the code with a for loop and a conditional operator (if-else) to create a column that is added to the data set (data frame, df). The code below creates this initial data set.
# col1 <- runif (12^5, 0, 2) col2 <- rnorm (12^5, 0, 2) col3 <- rpois (12^5, 3) col4 <- rchisq (12^5, 2) df <- data.frame (col1, col2, col3, col4) In the first part : vectorization, only true conditions, ifelse.
In this part: which, apply, byte-by-byte compilation, Rcpp, data.table, results.
')
Using which ()
Using the
which() command to select strings, you can achieve one-third the speed of Rcpp . # system.time({ want = which(rowSums(df) > 4) output = rep("less than 4", times = nrow(df)) output[want] = "greater than 4" }) # = 3 () user system elapsed 0.396 0.074 0.481 Use the apply family of functions instead of for loops
We use the
apply() function to implement the same logic and compare it with the vectorized for loop. The results grow with an increase in the number of orders, but they are slower than ifelse() and the version where the check was done outside the loop. This may be useful, but it may need some ingenuity for complex business logic. # apply system.time({ myfunc <- function(x) { if ((x['col1'] + x['col2'] + x['col3'] + x['col4']) > 4) { "greater_than_4" } else { "lesser_than_4" } } output <- apply(df[, c(1:4)], 1, FUN=myfunc) # 'myfunc' df$output <- output }) 
Using apply and for loops in R
Use byte compilations for the cmpfun () functions from the compiler package instead of the actual function.
This is probably not the best example to illustrate the effectiveness of byte-based compilation, since the resulting time is slightly higher than the usual form. However, for more complex functions, byte compilation has proven to be effective. I think it is worth trying on occasion.
# library(compiler) myFuncCmp <- cmpfun(myfunc) system.time({ output <- apply(df[, c (1:4)], 1, FUN=myFuncCmp) }) 
Apply, for loop and byte code compilation
Use Rcpp
Let's go to a new level. Before that, we increased speed and performance with various strategies and found that using
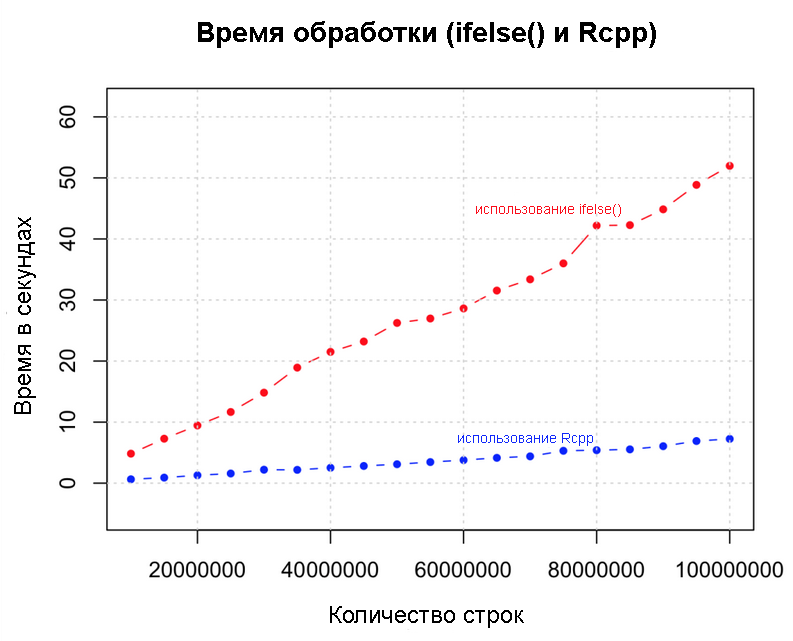
ifelse() most effective. What if we add another zero? Below we implement the same logic with Rcpp , with a data set of 100 million rows. We compare Rcpp and ifelse() speeds. library(Rcpp) sourceCpp("MyFunc.cpp") system.time (output <- myFunc(df)) # Rcpp Below is the same logic implemented in C ++ using the Rcpp package. Save the code below as “MyFunc.cpp” in your working session R session directory (or you will have to use sourceCpp using the full path). Note that the comment
// [[Rcpp::export]] required and must be placed immediately before the function you want to execute from R. // MyFunc.cpp #include using namespace Rcpp; // [[Rcpp::export]] CharacterVector myFunc(DataFrame x) { NumericVector col1 = as(x["col1"]); NumericVector col2 = as(x["col2"]); NumericVector col3 = as(x["col3"]); NumericVector col4 = as(x["col4"]); int n = col1.size(); CharacterVector out(n); for (int i=0; i 4){ out[i] = "greater_than_4"; } else { out[i] = "lesser_than_4"; } } return out; } 
Rcpp and ifelse performanceUse parallel processing if you have a multi-core computer.
Parallel processing:
# library(foreach) library(doSNOW) cl <- makeCluster(4, type="SOCK") # for 4 cores machine registerDoSNOW (cl) condition <- (df$col1 + df$col2 + df$col3 + df$col4) > 4 # system.time({ output <- foreach(i = 1:nrow(df), .combine=c) %dopar% { if (condition[i]) { return("greater_than_4") } else { return("lesser_than_4") } } }) df$output <- output Delete variables and clear memory as early as possible.
Remove more unnecessary objects in the code with
rm() as early as possible, especially before long cycles. Sometimes using gc() at the end of each iteration can help.Use less memory data structures
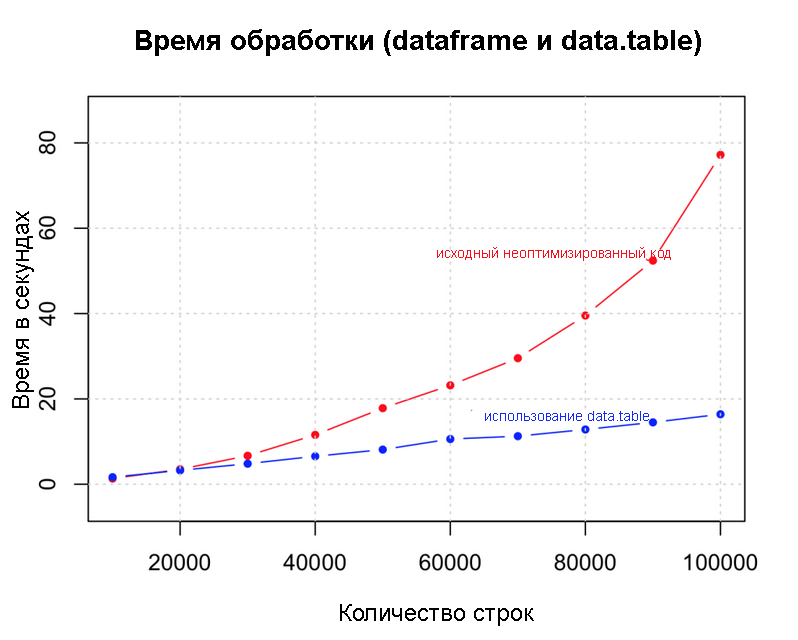
Data.table() is a great example because it does not overload the memory. This allows you to speed up operations like data fusion. dt <- data.table(df) # data.table system.time({ for (i in 1:nrow (dt)) { if ((dt[i, col1] + dt[i, col2] + dt[i, col3] + dt[i, col4]) > 4) { dt[i, col5:="greater_than_4"] # 5- } else { dt[i, col5:="lesser_than_4"] # 5- } } }) 
Dataframe and data.table
Speed: Results
Method: Speed, number of lines in df / elapsed time = n lines per second
Source: 1X, 120000 / 140.15 = 856.2255 rows per second (normalized to 1)
Vectorized: 738X, 120000 / 0.19 = 631578.9 lines per second
Only true conditions: 1002X, 120000 / 0.14 = 857142.9 lines per second
ifelse: 1752X, 1200000 / 0.78 = 1,500,000 lines per second
which: 8806X, 2985984 / 0.396 = 7540364 lines per second
Rcpp: 13476X, 1200000 / 0.09 = 11538462 lines per second
The numbers above are approximate and based on random runs. There are no calculations of the results for
data.table() , byte-by-code compilation and parallelization, since they will be very different in each particular case and depending on how you use them.Source: https://habr.com/ru/post/277693/
All Articles