DICOM Viewer from the inside. Functionality
Good day, habrasoobschestvo. I would like to continue to consider aspects of the implementation of DICOM Viewer, and today we will talk about the functionality.
So let's go.
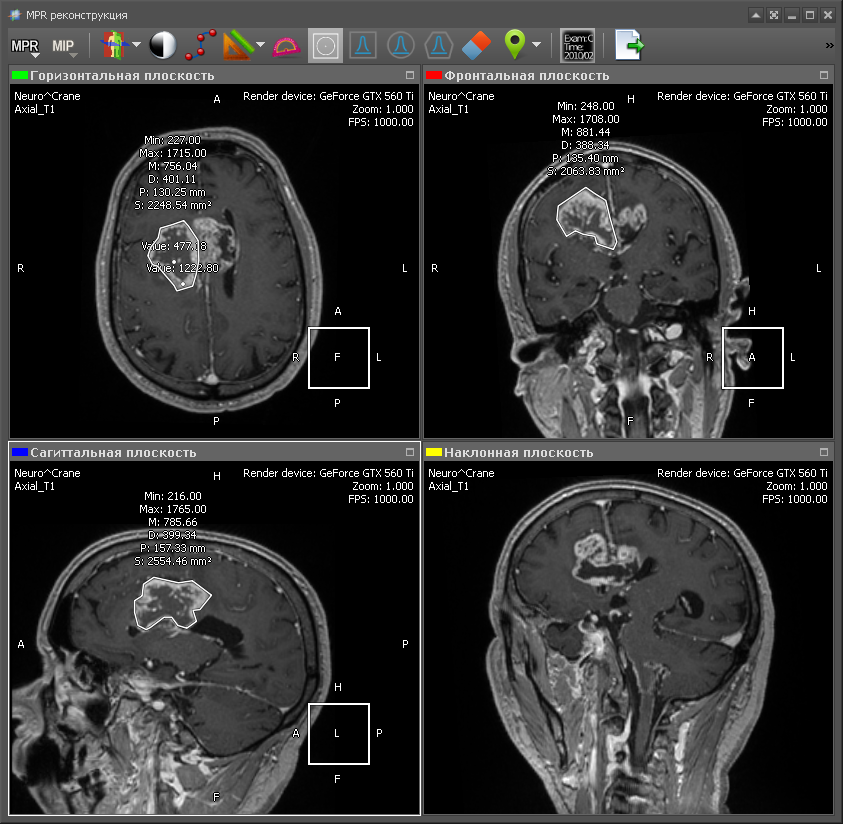
Multiplanar reconstruction allows you to create images from the original plane to the axial, frontal, sagittal or arbitrary plane. In order to build MPR, it is necessary to build a 3D volume model and “cut” it in the required planes. As a rule, the best quality of the MPR is obtained with computed tomography (CT), because in the case of CT, you can create a 3D model with a resolution that is the same in all planes. Therefore, the output MPR is obtained with the same resolution as the original images obtained from CT. Although there are MRI with good resolution. Here is an example of multiplanar reconstruction:
Green - axial plane (upper left);
Red - frontal plane (top right);
Blue - sagittal plane (lower left);
Yellow - an arbitrary plane (lower right).
')
The position of the lower right image is determined by the yellow line in the side view (upper left). This is the image obtained by “cutting” the 3D model with an inclined plane. To obtain the density value at a specific point of the plane, trilinear interpolation is used.

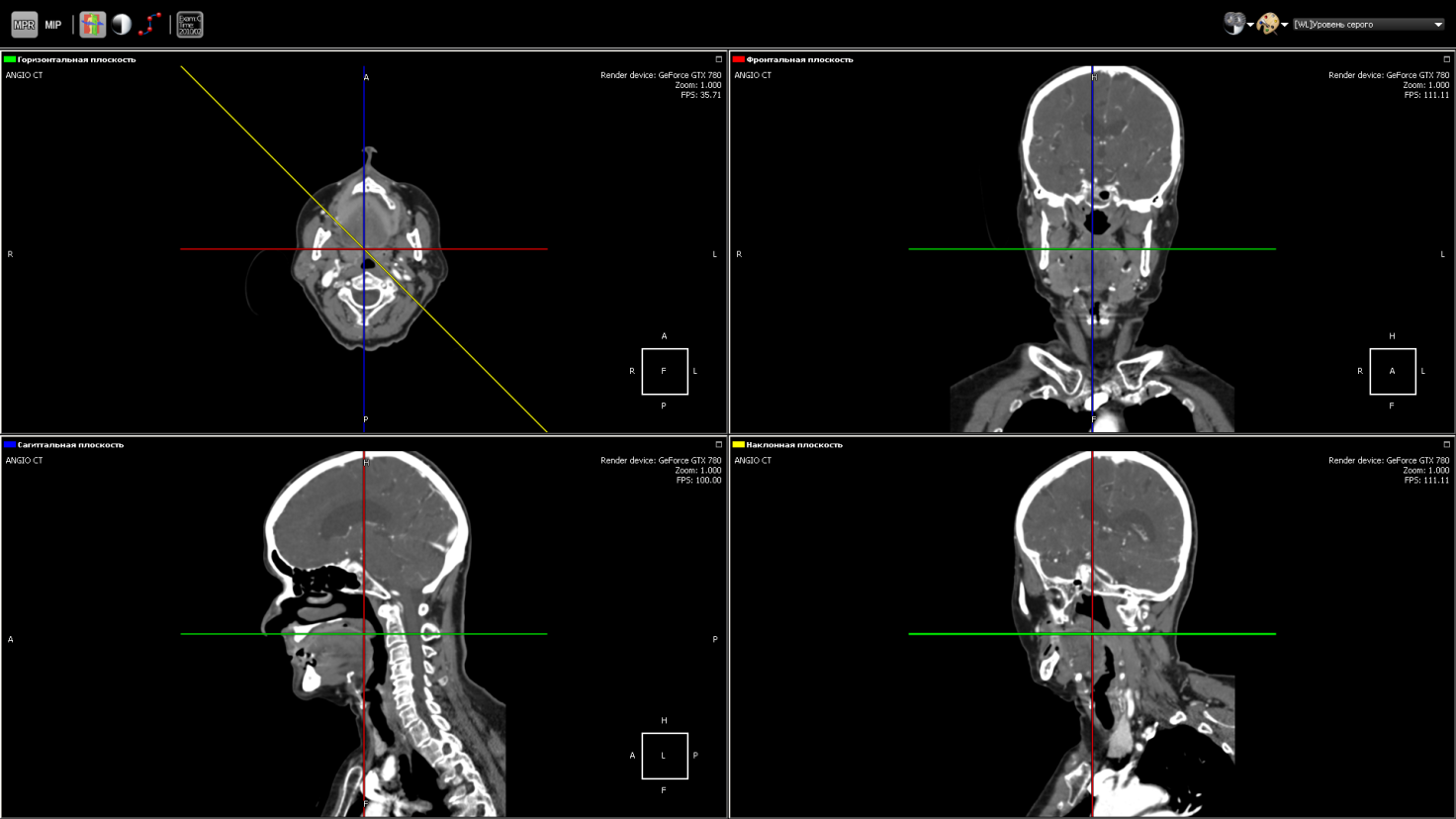
Same as MPR, but instead of an arbitrary plane you can take a curve, as shown in the figure. Used, for example, in dentistry for a panoramic picture of the teeth.
Each point on the curve defines the starting point of the trace, and the normal to the curve at this point corresponds to the direction of the Y-axis in the two-dimensional image for this point. The X axis of the image corresponds to the curve itself. That is, at each point of the two-dimensional image, the direction of the X axis is the tangent to the curve at the corresponding point on the curve.
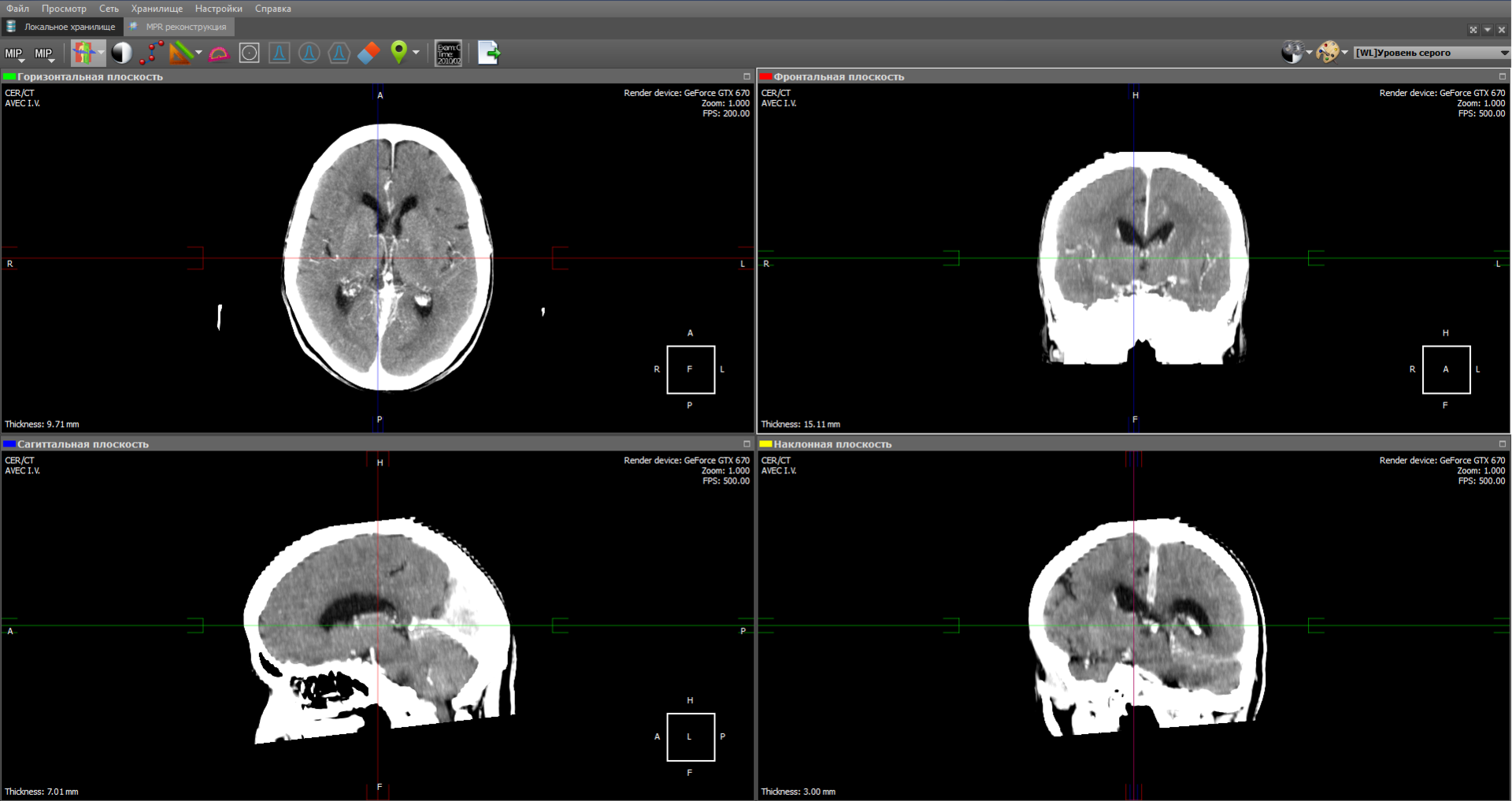
The minimum intensity values indicate soft tissue. Whereas the values of maximum intensity correspond to the brightest areas of a three-dimensional object — these are either the most dense tissues or organs saturated with contrast material. The minimum / average / maximum intensity value is taken in the range (as shown in the figure by the dotted lines). The minimum value throughout the model will take the air.
The MIP calculation algorithm is very simple: choose a plane on the 3D model - let it be the XY plane. Then we go along the Z axis and select the maximum intensity value on the specified range and display it on the 2D plane:
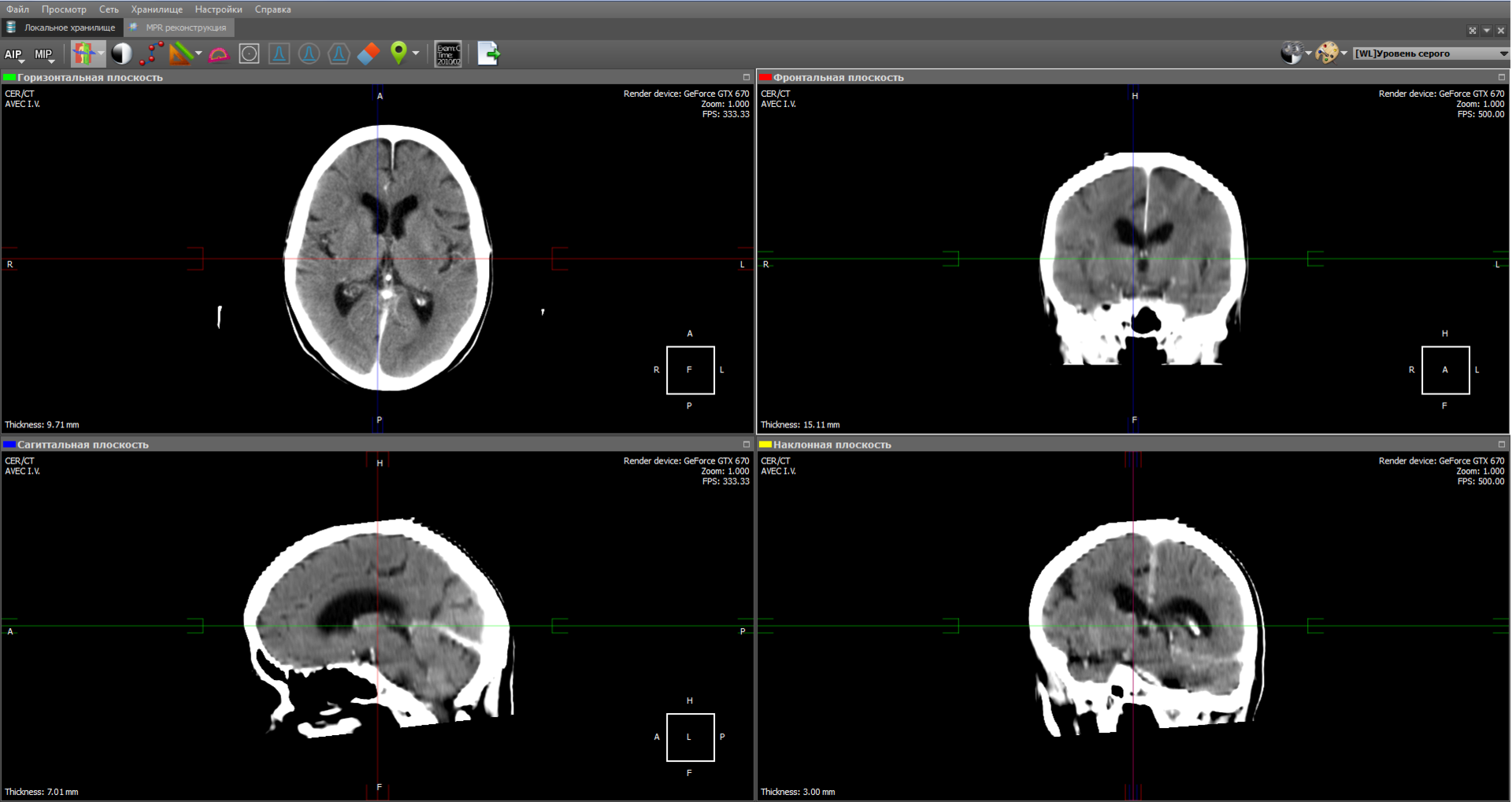
The image obtained by the projection of medium intensity is close to the usual X-ray image:
Some types of radiological studies do not give the desired effect without the use of a contrast agent, because they do not reflect certain types of tissues and organs. This is due to the fact that in the human body there are tissues whose density is about the same. To distinguish these tissues from each other, use a contrast agent, which gives the blood more intensity. Also, a contrast agent is used to visualize the vessels during angiography.
Angiography is a technique that allows visualization of the bloodstream systems (veins and vessels) of various organs. For this purpose, a contrast agent is used, which is injected into the test organ, and an X-ray machine, which creates images during the injection of a contrast agent. Thus, at the output of the apparatus, a set of images with different degrees of visualization of blood flow is obtained:
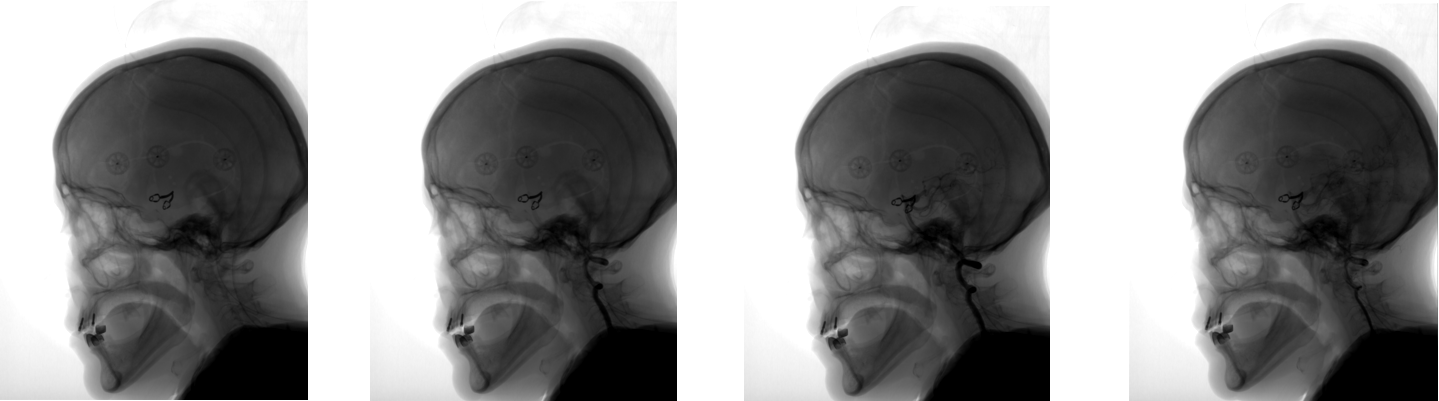
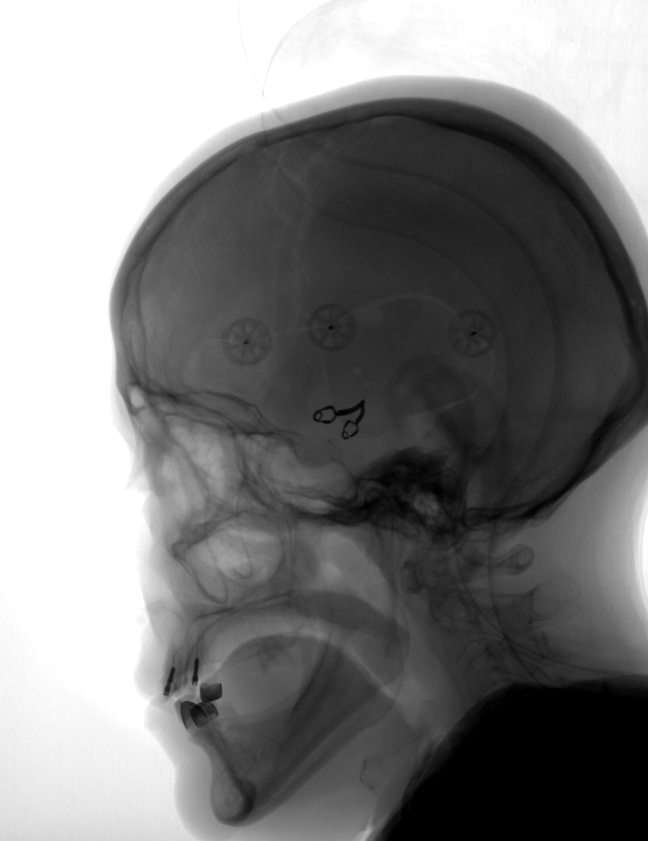
However, along with the veins and vessels in the pictures visible tissue of other organs, such as the skull. DSA (Digital subtraction angiography) mode allows you to visualize only the blood flow without any other tissue. How it works? We take the image of a series in which the blood flow has not yet been visualized by a contrast agent. As a rule, this is the first image of the series, the so-called mask:
Then we subtract this image from all other images in the series. We get the following image:
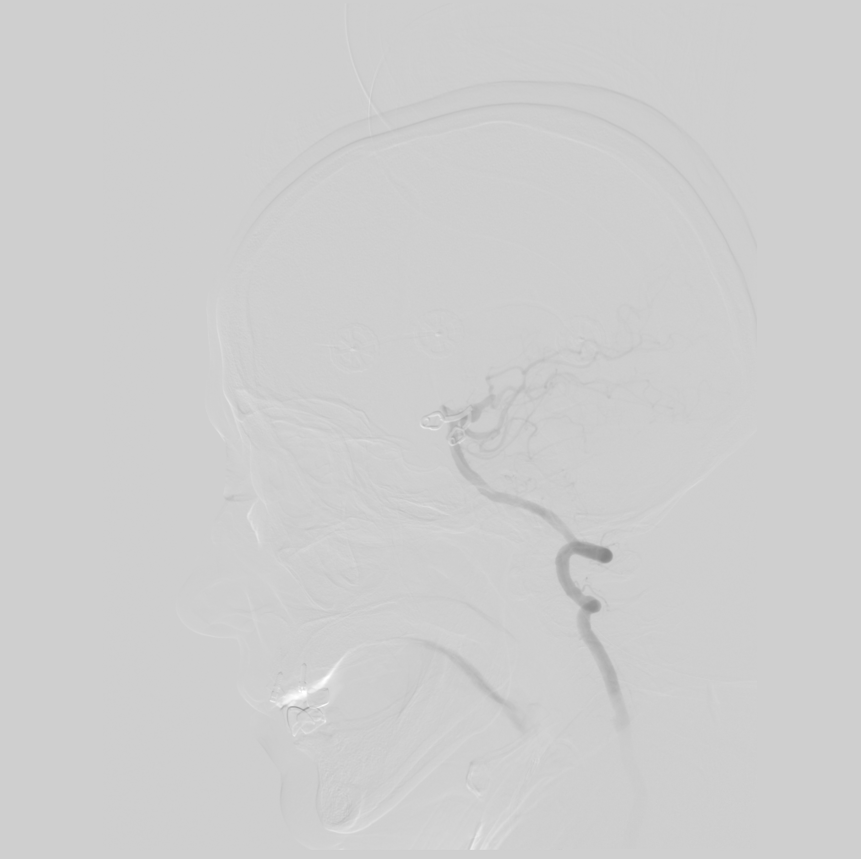
In this image, blood flow is clearly visible and almost no other tissue is visible, which allows for more accurate diagnosis.
The Clipping Box tool allows you to see the bones and anatomical tissues in the section, as well as show the internal organs from the inside. The tool is implemented at the renderer level, simply limiting the area of raytracing.
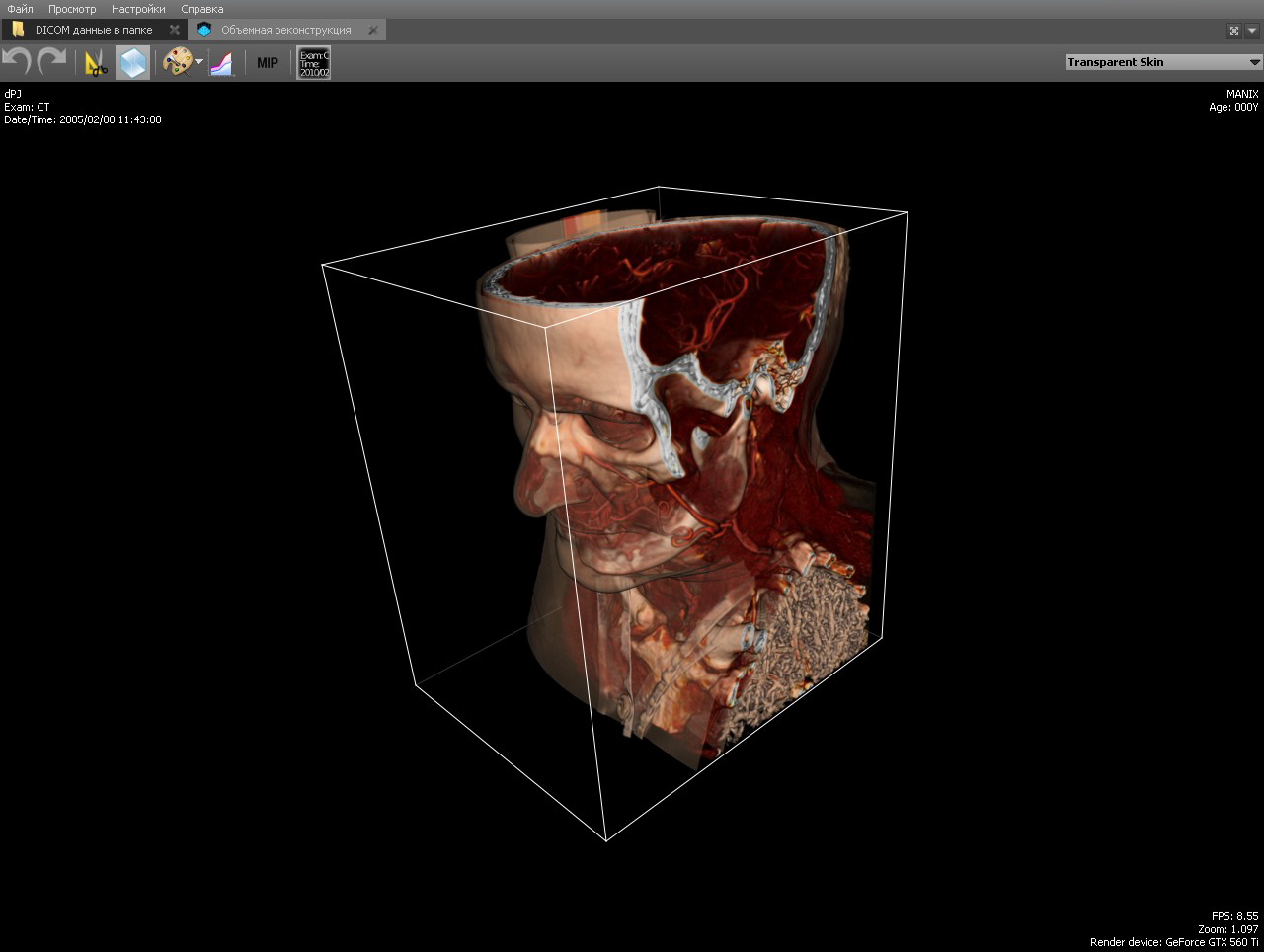
In the implementation of the region of raytracing is limited to planes with normals directed in the direction of clipping. That is, the cube is represented by six planes.
The tool is similar to the previous one and allows you to delete a volume fragment under an arbitrary polygon:
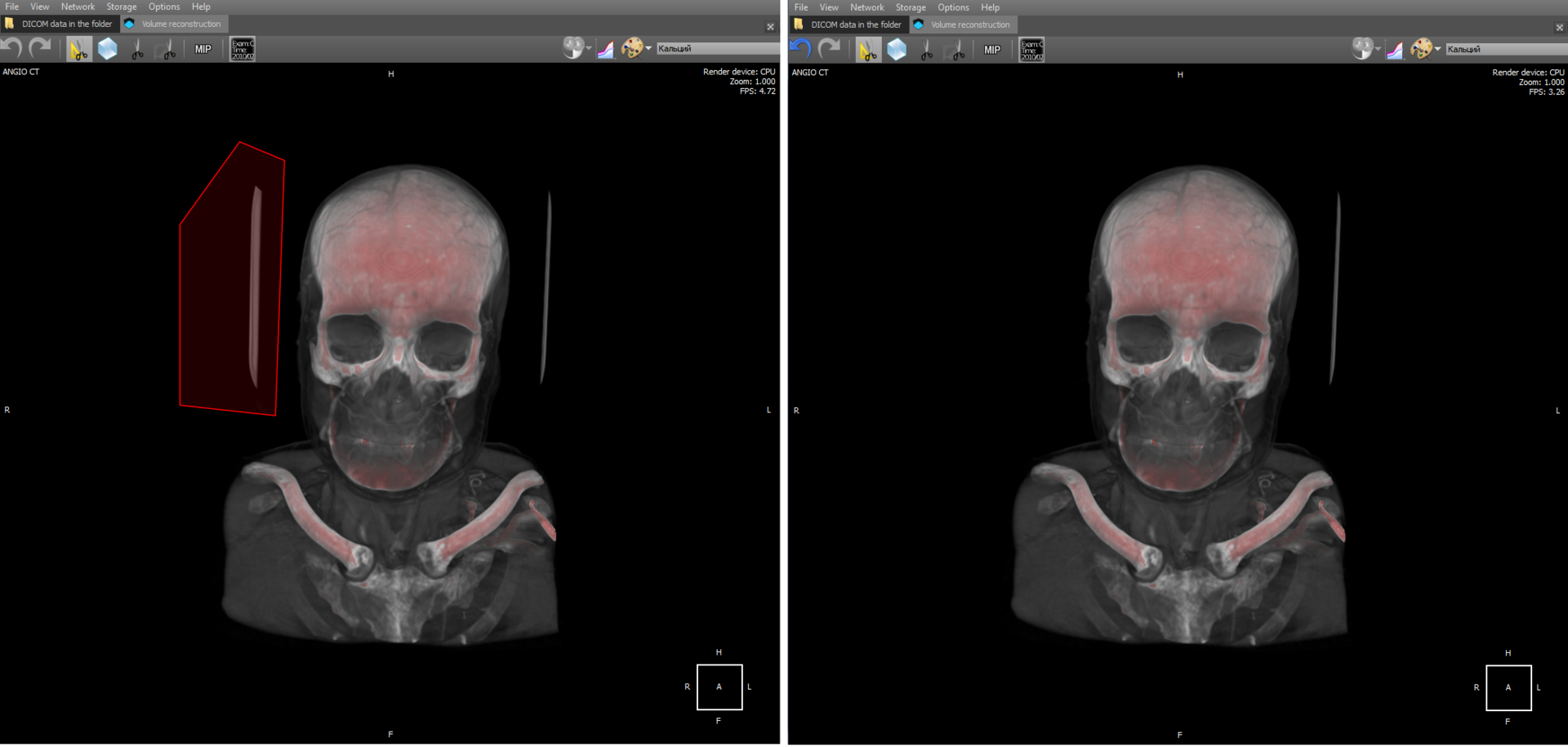
By cutting, one should understand voxel vanishing in a 3D model trapped in a polygon region.
Also there is a tool "Scissors", which allow you to remove parts of the 3D-model on the principle of connectivity. Implementation: when an object is selected, a cyclic search for nearby connected voxels occurs until all nearby voxels are scanned. Then all scanned voxels are deleted.
In 3D, organs can be measured at any angle, which is impossible in some cases in 2D.
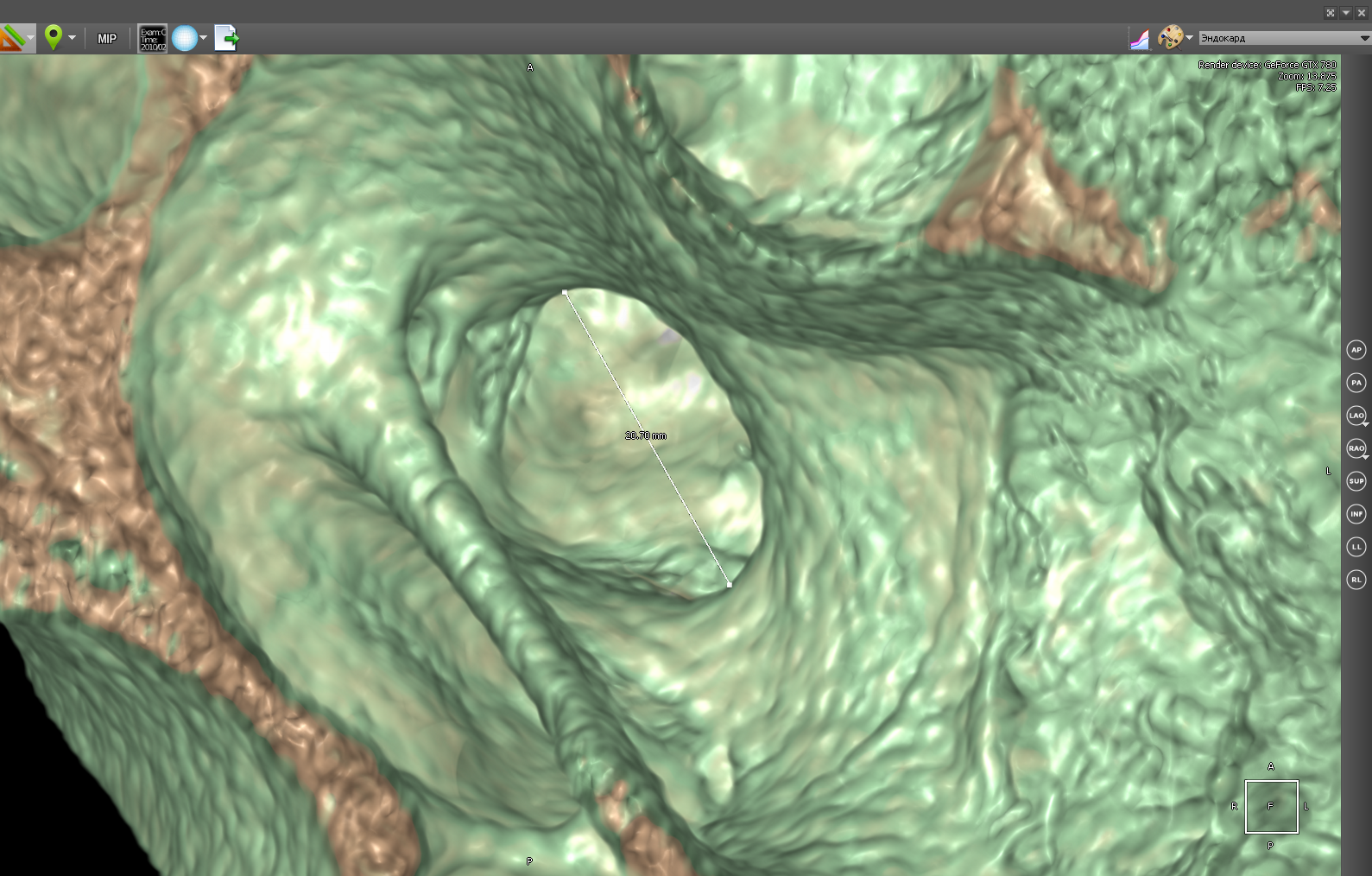
In 3D mode, you can also use a polygonal ruler:
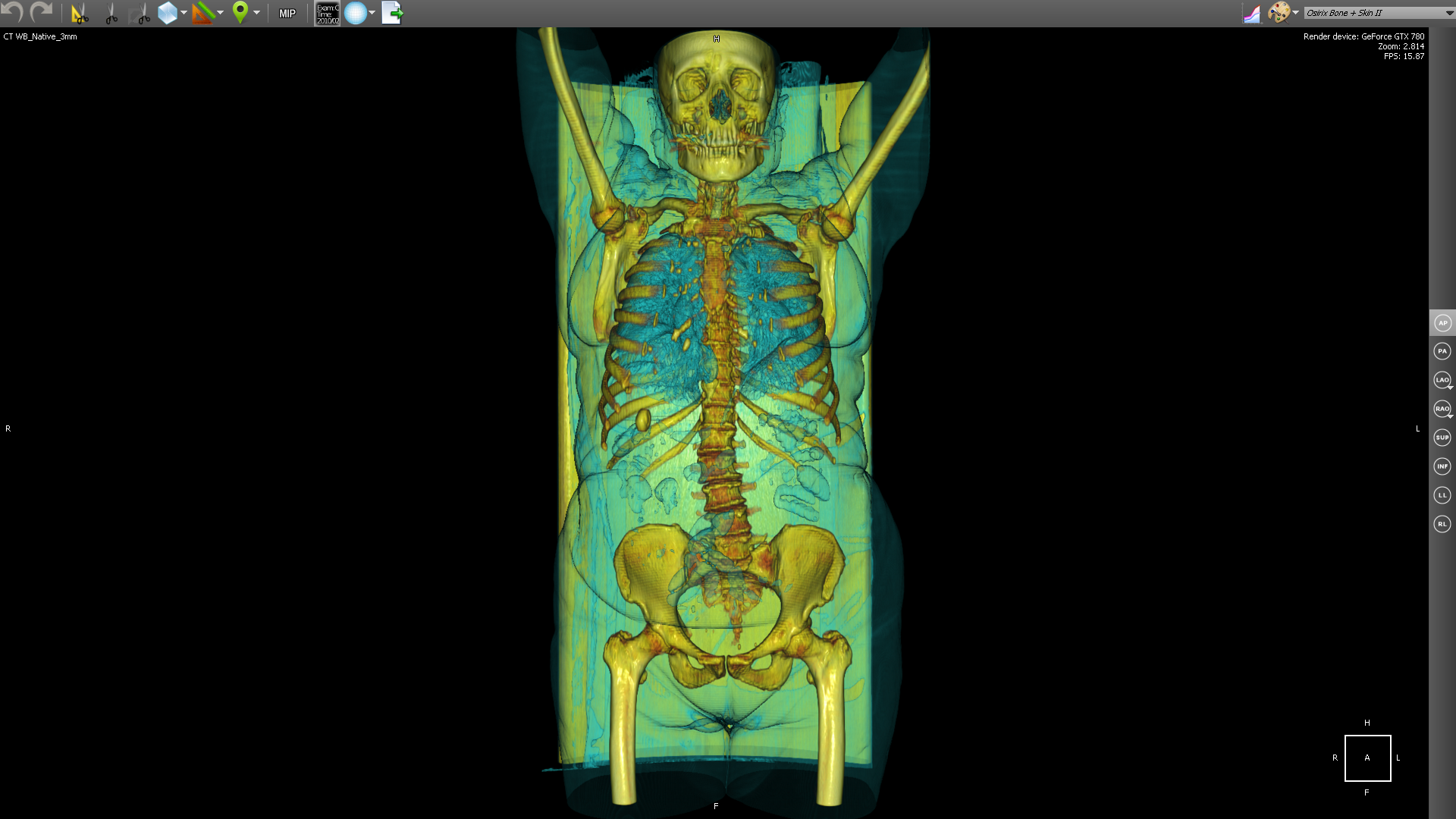
PET-CT (eng. PET-CT) is a relatively new technology, which is a research method of nuclear medicine. Is a method of multimodal tomography. The fourth dimension in this case is modality (PET and CT). Intended mainly for detecting cancerous tumors.
CT helps to get the anatomical structure of the human body:
and PET shows certain areas of concentration of a radioactive substance, which is directly related to the intensity of blood supply to a given area.
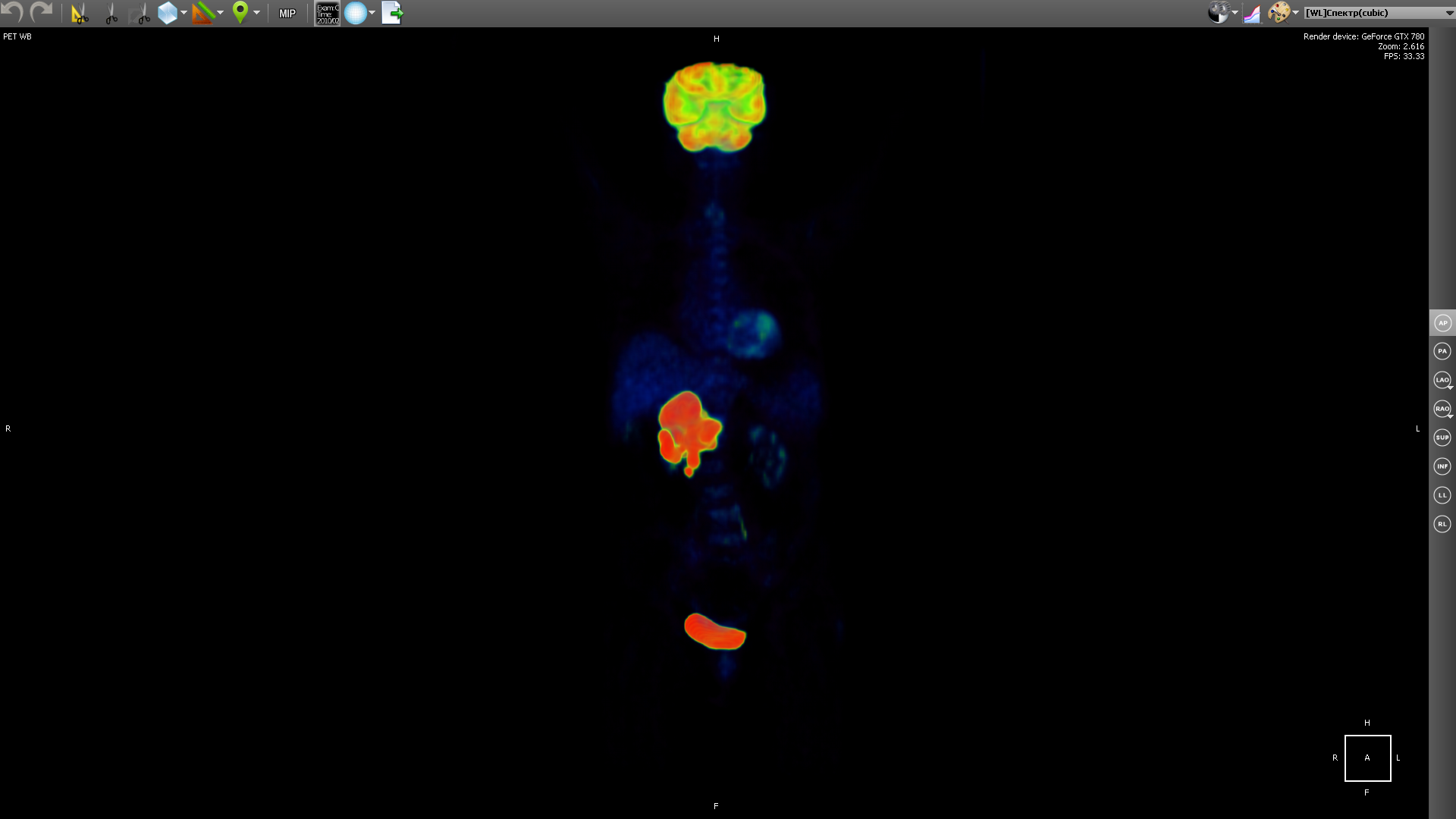
PET gets a picture of biochemical activity by detecting radioactive isotopes in the human body. The radioactive substance accumulates in organs saturated with blood. Then the radioactive substance undergoes positron beta decay. The resulting positrons are further annihilated with electrons from the surrounding tissue, resulting in the emission of gamma-ray pairs, which are detected by the device, and then a 3D image is constructed based on the information obtained.
The selection of a radioactive isotope determines the biological process that one wishes to track in the research process. The process may be metabolism, transport of substances, etc. Behavior of the process, in turn, is the key to the correct diagnosis of the disease. The image above shows a tumor in the liver area.
But based on PET, it is difficult to understand in which part of the body the area with the maximum concentration of the radioactive substance is located. By combining body geometry (CT) and areas saturated with blood with a high concentration of radioactive substance (PET), we get:
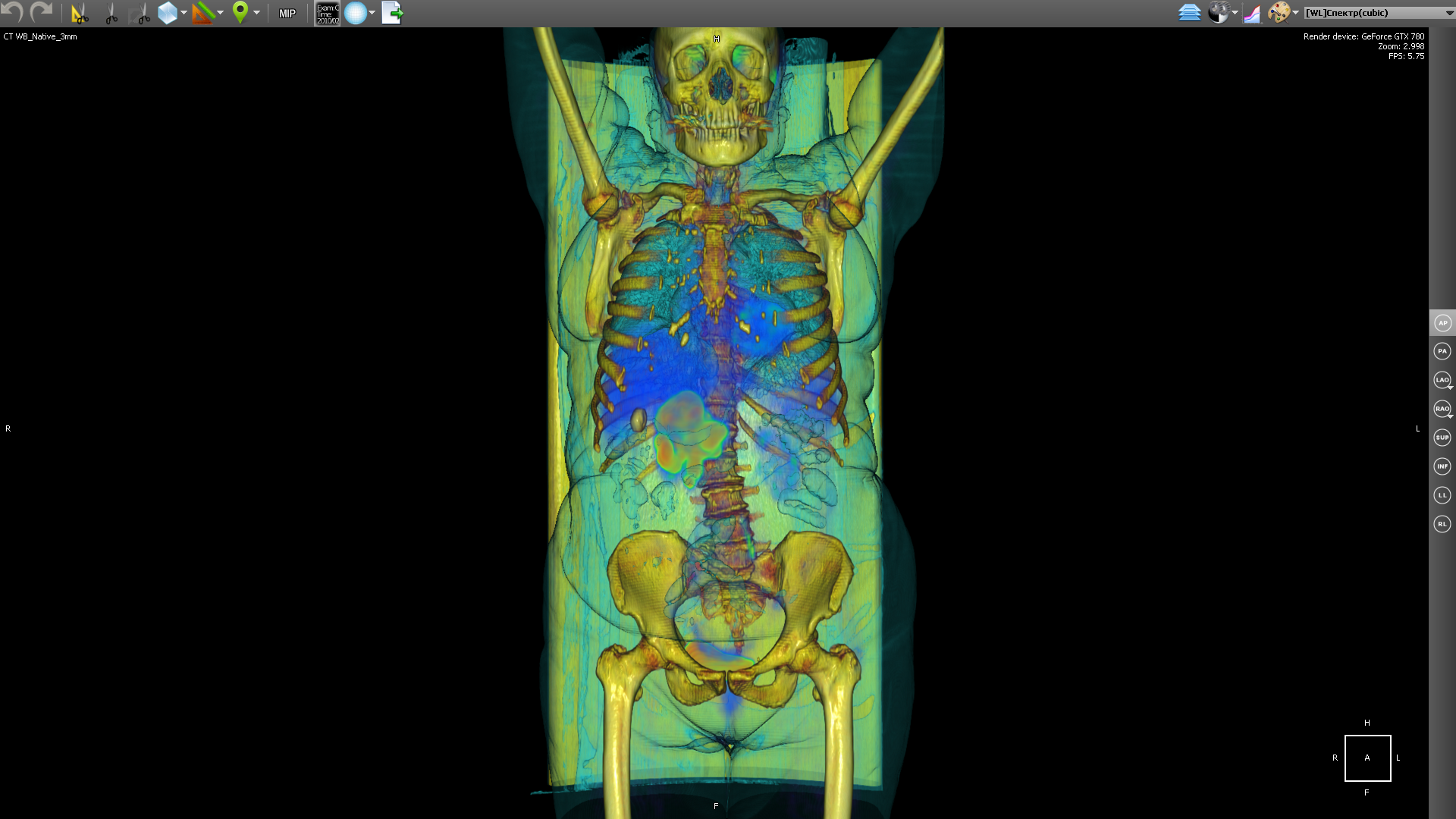
Radioactive isotopes with different half-lives are used as a radioactive substance for PET. Fluorine-18 (fluorodeoxyglucose) is used to form all kinds of malignant tumors, iodine-124 is used to diagnose thyroid cancer, and gallium-68 is used to detect neuroendocrine tumors.
The Fusion functionality forms a new series in which the images of both modalities (both PET and CT) are combined. In the implementation, the images of both modalities are mixed, and then sorted along the Z axis (we assume that X and Y are the image axes). In fact, it turns out that the images in the series alternate (PET, CT, PET, CT ...). This series is further used to draw 2D fusion and 3D fusion. In the case of 2D fusion, images are drawn in pairs (PET-CT) in ascending order of Z:
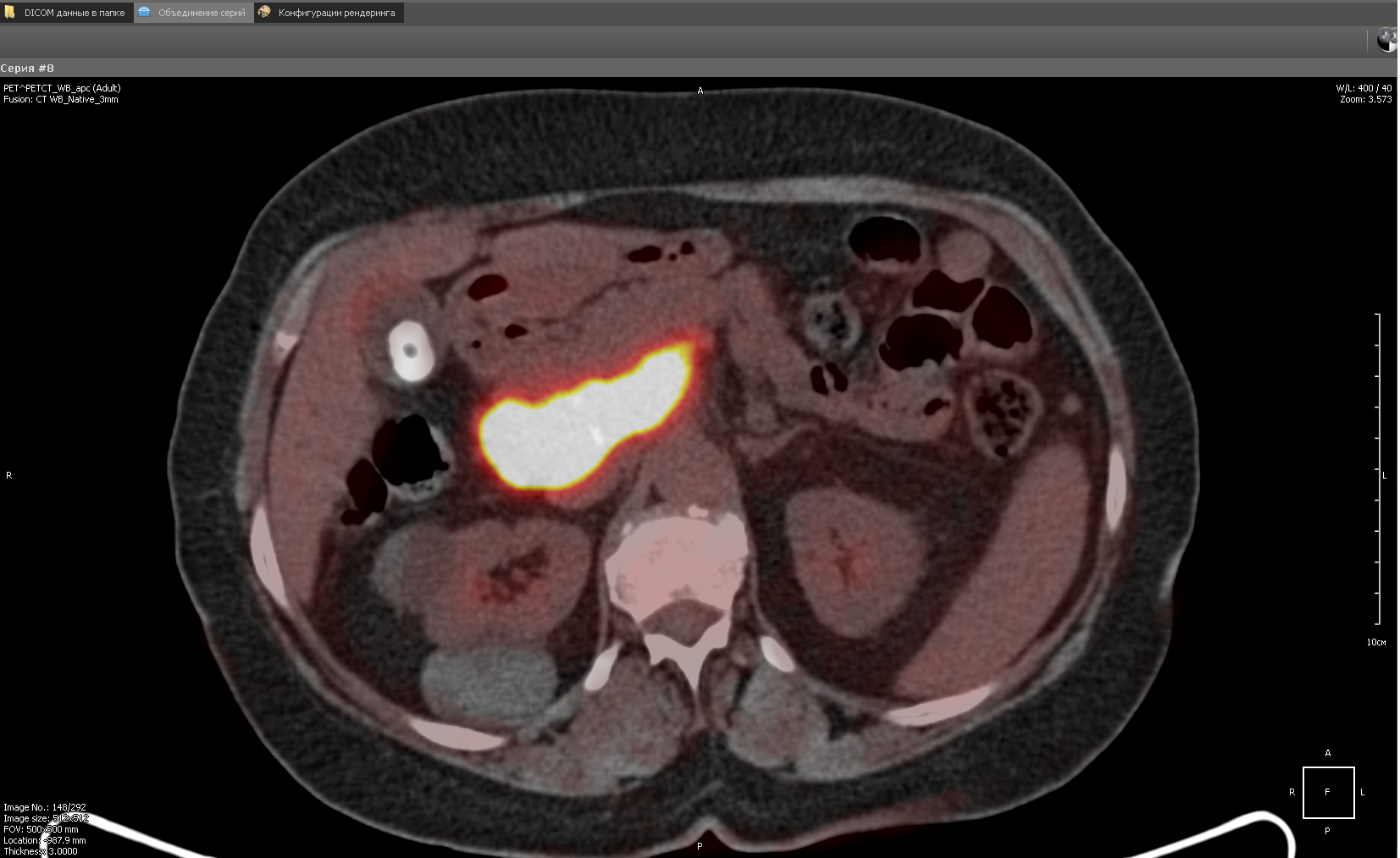
In this case, the CT image was first drawn, then the PET.
3D fusion is implemented for a video card on CUDA. Both 3D-models - PET and CT are drawn on the video card at the same time and a real multimodal fusion is obtained. On the processor, the fusion also works, but it works a little differently. The fact is that on the processor both models are represented in memory as separate octo-trees. Therefore, when drawing it is necessary to trace two trees and synchronize the skipping of transparent voxels. And this would significantly reduce the speed of work. Therefore, it was decided to simply overlay the result of rendering one 3D-model on top of another.
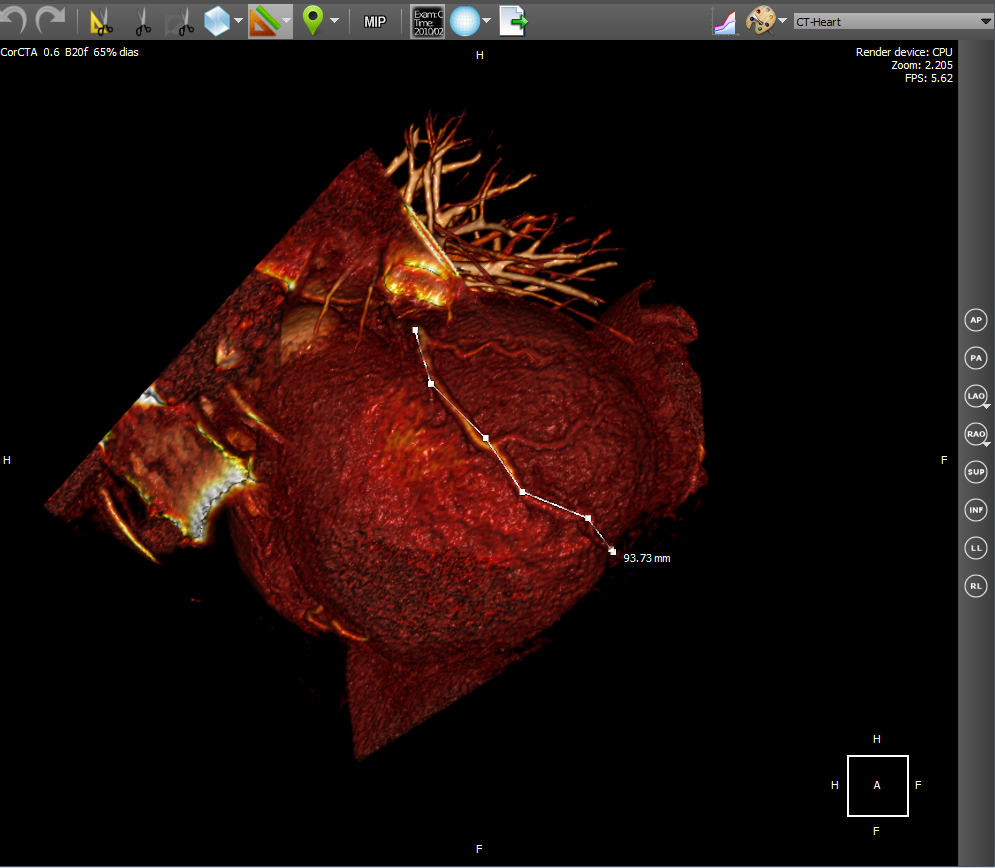
Cardiac CT technology is used to diagnose various disorders of the heart, including coronary heart disease, pulmonary thromboembolism and other diseases.
4D Cardiac CT is a 3D in time. Those. It turns out a small video, which we will call a film loop, in which each frame will be a 3D object. The source data is a set of dicom-images for all frames of a film loop at once. In order to convert a set of images into a film loop, you must first group the source images into frames, and then create 3D for each frame. Building a 3D object at the frame level is the same as for any series of dicom images. We use heuristic image sorting to group by frame, using the position of the image on the Z axis (assuming that X and Y are image axes). We believe that after grouping by frames, the same number of images is obtained in each frame. Switching the frame actually comes down to switching the 3D model.
5D Fusion Pet - CardiacCT is a 4D Cardiac CT with the addition of fusion with PET as the fifth dimension. In the implementation, we first create two film loops: with CardiacCT and with PET. Then we make fuision of the corresponding frames of the movie band, which gives us a separate series. Then we build the resulting 3D series. It looks like this:
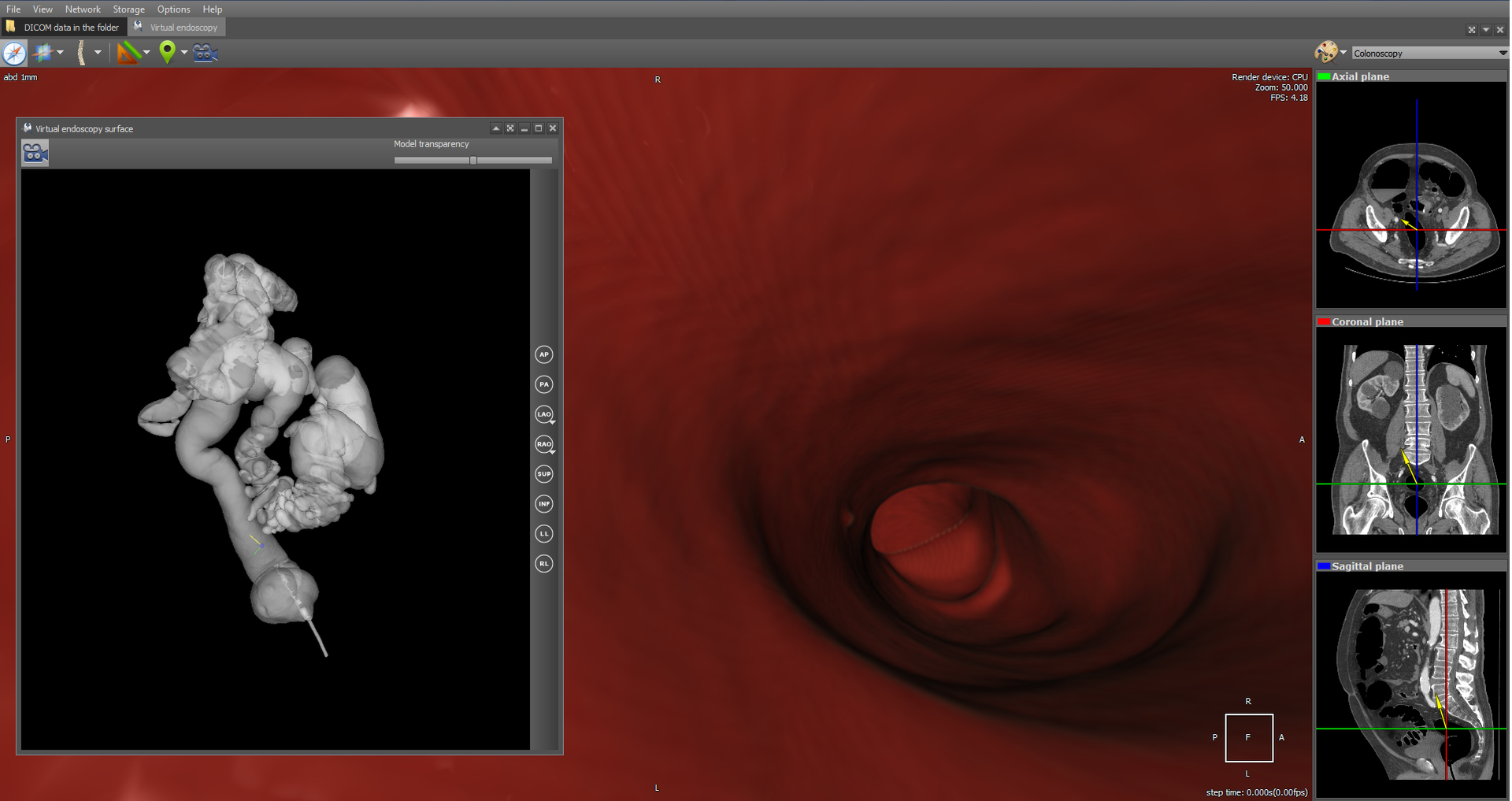
As an example of virtual endoscopy, we will consider virtual colonoscopy, since it is the most common type of virtual endoscopy. Virtual colonoscopy allows to construct a volumetric reconstruction of the abdominal cavity area on the basis of CT data and to make a diagnosis using this 3D reconstruction. In the viewer there is a fly-through camera tool with MPR navigation:
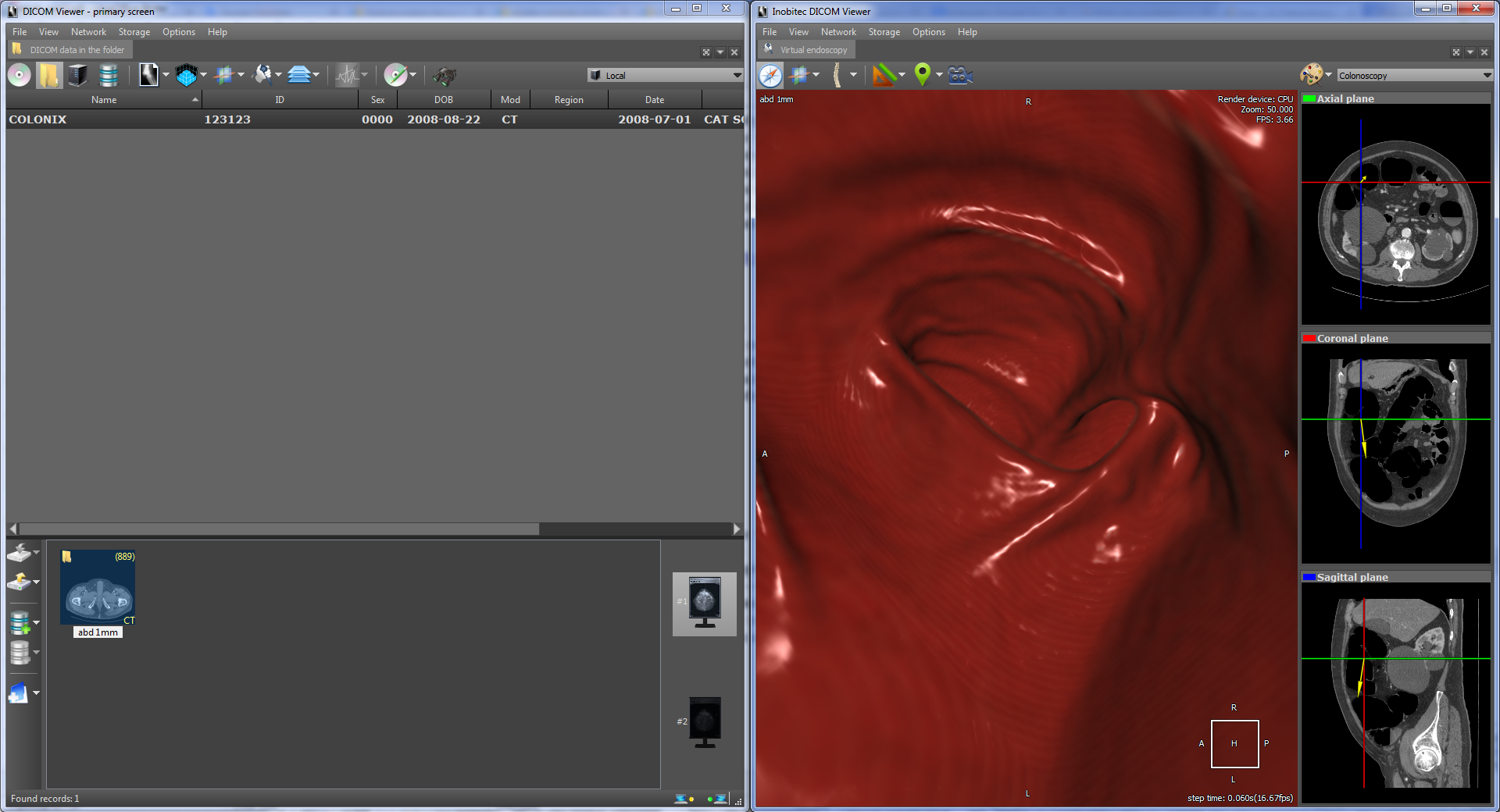
which also allows you to automatically follow the anatomical structure. In particular, it allows you to view the intraintestinal region automatically. Here's what it looks like:
The flight of the camera represents a series of consecutive movements in the intraintestinal region. For each step, the vector of camera movement to the next part of the anatomical structure is calculated. The calculation is based on transparent voxels in the next part of the anatomical structure. In fact, a certain voxel is calculated among transparent ones. The initial displacement vector is defined by the camera vector. In the Camera Flight tool, an exclusively perspective projection is used
There is also a functional for automatic intestinal segmentation, i.e. functionality to separate the intestinal region from the rest of the anatomy:
It is also possible to navigate through a segmented 3D model (the Show camera orientation button), which, by a mouse click on the 3D model, moves the camera to the appropriate position in the original anatomy.
Segmentation is implemented using a wave algorithm . It is believed that the anatomy is closed in the sense that it is not in contact with other organs and external space.
A separate module in the viewer implements data reading from the Waveform and its drawing. DICOM ECG Waveform is a special format for storing data from electrocardiogram leads defined by the DICOM standard. These electrocardiograms are twelve leads - 3 standard, 3 reinforced and 6 chest. The data of each lead is a sequence of measurements of the electrical voltage on the surface of the body. In order to draw the voltage, you need to know the scale vertically in mm / mV and the horizontal scale in mm / s:
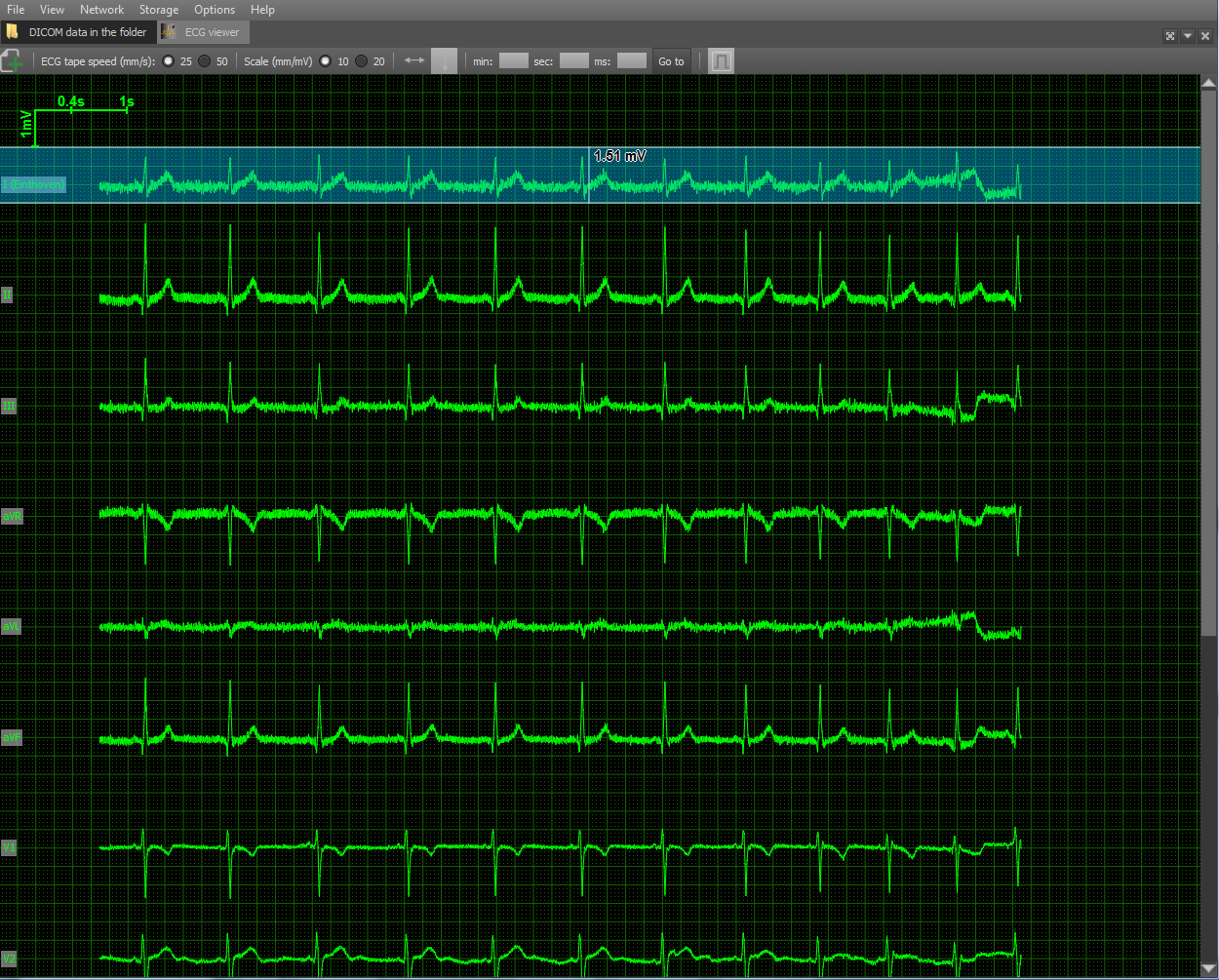
As auxiliary attributes, the grid is also drawn for ease of measuring distances and a scale in the upper left corner. Scale options are selected taking into account medical practice: 10 and 20 mm / mV vertically, 25 and 50 mm / sec horizontally. Also implemented tools for measuring the distance horizontally and vertically.
DICOM-Viewer, among other things, is a full-fledged DICOM client. It is possible to search on the PACS server, retrieve data from it, etc. The functions of the DICOM client are implemented using the open DCMTK library. Consider a typical use-case of the DICOM client on the example of the viewer. Perform a search for stages on a remote PACS server:
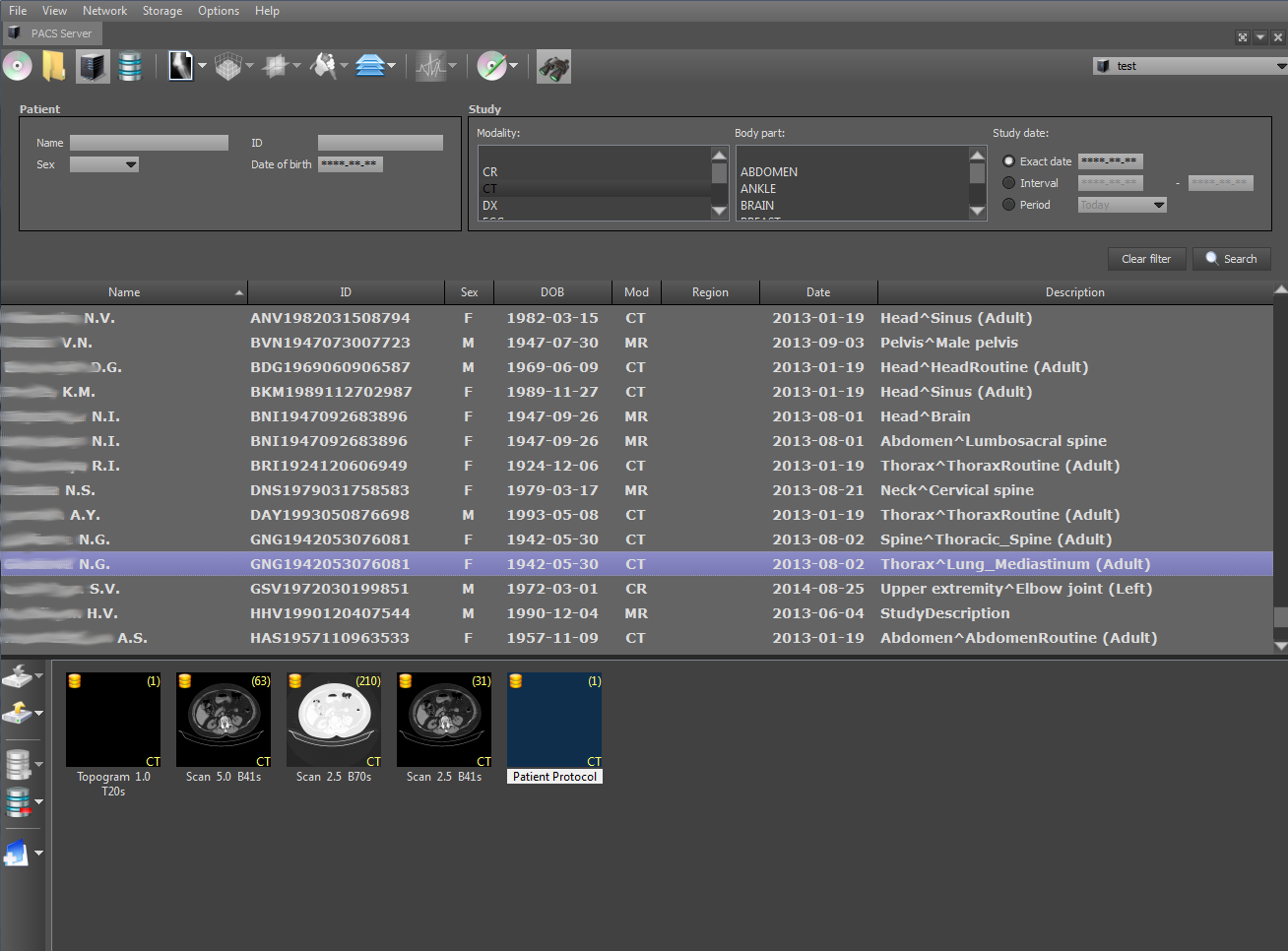
When a stage is selected, the series for the selected stage and the number of images in them are displayed at the bottom. The PACS server on which the search will be performed is indicated on the top right. The search can be parameterized by refining the search criteria: PID, study date, patient name, etc. The client search is performed by the C-FIND SCU command using the DCMTK library, which operates at one of the levels: STUDY, SERIES and IMAGE.
Next, images of the selected series can be downloaded using the C-GET-SCU and C-MOVE-SCU commands. The DICOM protocol obliges the parties to the connection, i.e. client and server, agree in advance what type of data they are going to transfer through this connection. A data type is a combination of the values of the SOPClassUID and TransferSyntax parameters. SOPClassUID determines the type of operation that is scheduled to be performed through this connection. The most commonly used SOPClassUIDs are: Verification SOP Class (ping server), Storage Service Class (image saving), Printer Sop Class (printing on a DICOM printer), CT Image Storage (saving CT images), MR Image Storage (saving image MRI) and others. TransferSyntax determines the format of a binary file. Popular TransferSyntaxs: Little Endian Explicit, Big Endian Implicit, JPEG Lossless Nonhierarchical (Processes 14). That is, in order to transfer MRI images in the Little Endian Implicit format, then MR Image Storage - Little Endian Explicit must be added to the connection.
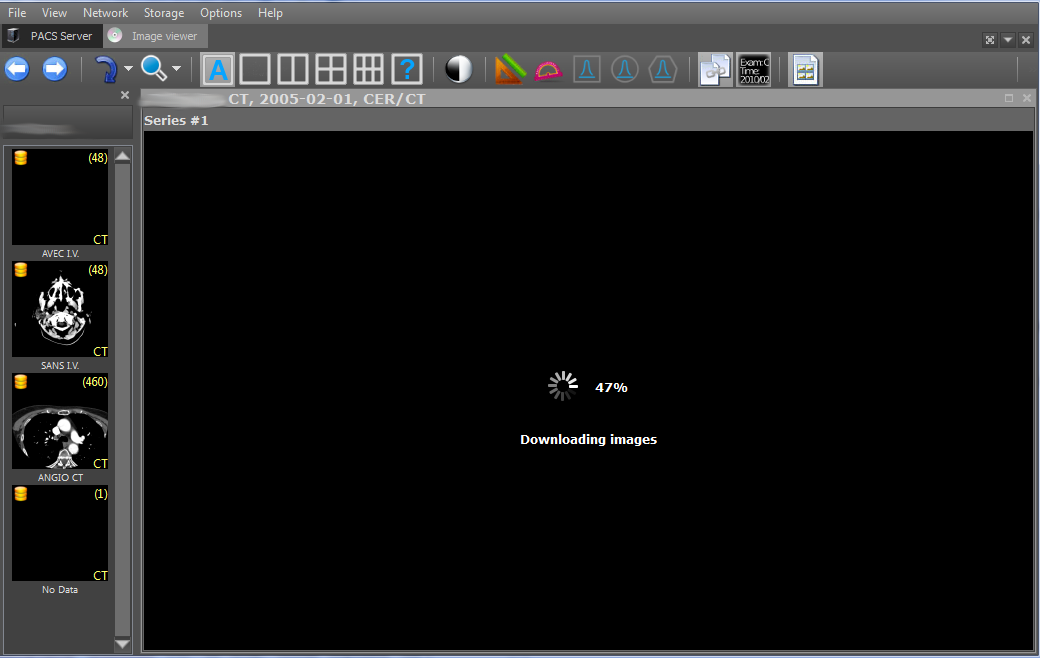
Downloaded images are saved to the local storage and, when re-viewed, are downloaded from it, which allows to increase the performance of the viewer. Saved series are marked with a yellow icon in the upper left corner of the first image of the series.
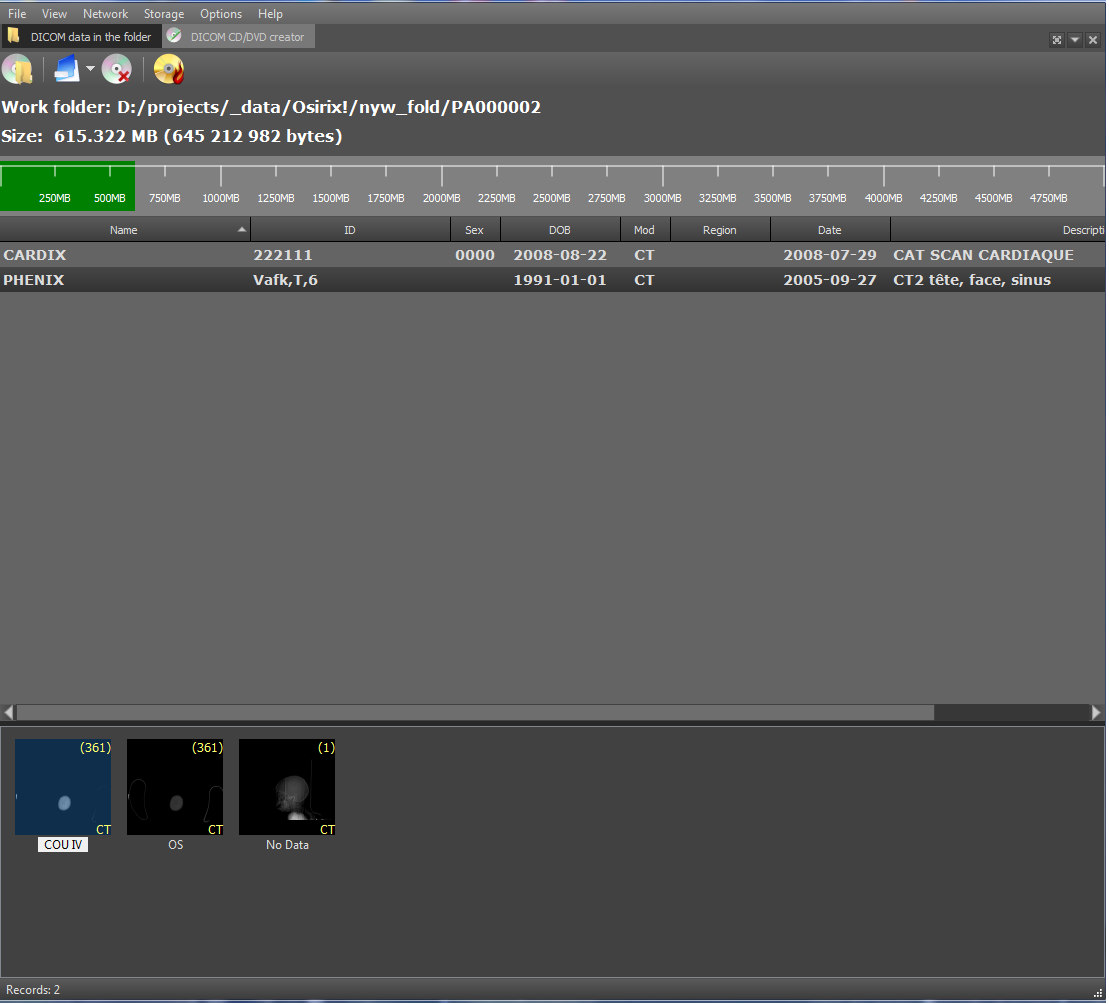
Also, DicomViewer as a DICOM client can burn discs with studies in the DICOMDIR format. The DICOMDIR format is implemented as a binary file that contains relative paths to all DICOM files that are written to disk. Implemented using the DCMTK library. When reading a disc, the paths to all files from DICOMDIR are read and then loaded. To add stages and series to DICOMDIR, the following interface was developed:
That's all that I wanted to tell you about the functionality of DicomViewer. As always, feedback from qualified professionals is very welcome.
Viewer Link:
DICOM Viewer x86
DICOM Viewer x64
Data examples:
MANIX - for common examples (MPR, 2D, 3D, etc.)
COLONIX - for virtual colonoscopy
FIVIX - 4D CARDIAC-CT
CEREBRIX - Fusion PET-CT

So let's go.
2D toolbox
Multiplanar Reconstruction (MPR)
Multiplanar reconstruction allows you to create images from the original plane to the axial, frontal, sagittal or arbitrary plane. In order to build MPR, it is necessary to build a 3D volume model and “cut” it in the required planes. As a rule, the best quality of the MPR is obtained with computed tomography (CT), because in the case of CT, you can create a 3D model with a resolution that is the same in all planes. Therefore, the output MPR is obtained with the same resolution as the original images obtained from CT. Although there are MRI with good resolution. Here is an example of multiplanar reconstruction:

Green - axial plane (upper left);
Red - frontal plane (top right);
Blue - sagittal plane (lower left);
Yellow - an arbitrary plane (lower right).
')
The position of the lower right image is determined by the yellow line in the side view (upper left). This is the image obtained by “cutting” the 3D model with an inclined plane. To obtain the density value at a specific point of the plane, trilinear interpolation is used.
Multiplanar reconstruction on an arbitrary curve (curved MPR)

Same as MPR, but instead of an arbitrary plane you can take a curve, as shown in the figure. Used, for example, in dentistry for a panoramic picture of the teeth.
Each point on the curve defines the starting point of the trace, and the normal to the curve at this point corresponds to the direction of the Y-axis in the two-dimensional image for this point. The X axis of the image corresponds to the curve itself. That is, at each point of the two-dimensional image, the direction of the X axis is the tangent to the curve at the corresponding point on the curve.
The projection of the minimum / average / maximum intensity (MIP)
The minimum intensity values indicate soft tissue. Whereas the values of maximum intensity correspond to the brightest areas of a three-dimensional object — these are either the most dense tissues or organs saturated with contrast material. The minimum / average / maximum intensity value is taken in the range (as shown in the figure by the dotted lines). The minimum value throughout the model will take the air.
The MIP calculation algorithm is very simple: choose a plane on the 3D model - let it be the XY plane. Then we go along the Z axis and select the maximum intensity value on the specified range and display it on the 2D plane:

The image obtained by the projection of medium intensity is close to the usual X-ray image:

Some types of radiological studies do not give the desired effect without the use of a contrast agent, because they do not reflect certain types of tissues and organs. This is due to the fact that in the human body there are tissues whose density is about the same. To distinguish these tissues from each other, use a contrast agent, which gives the blood more intensity. Also, a contrast agent is used to visualize the vessels during angiography.
DSA mode for angiography
Angiography is a technique that allows visualization of the bloodstream systems (veins and vessels) of various organs. For this purpose, a contrast agent is used, which is injected into the test organ, and an X-ray machine, which creates images during the injection of a contrast agent. Thus, at the output of the apparatus, a set of images with different degrees of visualization of blood flow is obtained:

However, along with the veins and vessels in the pictures visible tissue of other organs, such as the skull. DSA (Digital subtraction angiography) mode allows you to visualize only the blood flow without any other tissue. How it works? We take the image of a series in which the blood flow has not yet been visualized by a contrast agent. As a rule, this is the first image of the series, the so-called mask:

Then we subtract this image from all other images in the series. We get the following image:

In this image, blood flow is clearly visible and almost no other tissue is visible, which allows for more accurate diagnosis.
3D toolkit
Clipping Box tool
The Clipping Box tool allows you to see the bones and anatomical tissues in the section, as well as show the internal organs from the inside. The tool is implemented at the renderer level, simply limiting the area of raytracing.

In the implementation of the region of raytracing is limited to planes with normals directed in the direction of clipping. That is, the cube is represented by six planes.
Volume Editing Tools - Polygon Cut
The tool is similar to the previous one and allows you to delete a volume fragment under an arbitrary polygon:

By cutting, one should understand voxel vanishing in a 3D model trapped in a polygon region.
Also there is a tool "Scissors", which allow you to remove parts of the 3D-model on the principle of connectivity. Implementation: when an object is selected, a cyclic search for nearby connected voxels occurs until all nearby voxels are scanned. Then all scanned voxels are deleted.
Ruler in 3D
In 3D, organs can be measured at any angle, which is impossible in some cases in 2D.

In 3D mode, you can also use a polygonal ruler:

4d toolkit
Combination of several tomographic series in 3D (Fusion PET-CT)
PET-CT (eng. PET-CT) is a relatively new technology, which is a research method of nuclear medicine. Is a method of multimodal tomography. The fourth dimension in this case is modality (PET and CT). Intended mainly for detecting cancerous tumors.
CT helps to get the anatomical structure of the human body:

and PET shows certain areas of concentration of a radioactive substance, which is directly related to the intensity of blood supply to a given area.

PET gets a picture of biochemical activity by detecting radioactive isotopes in the human body. The radioactive substance accumulates in organs saturated with blood. Then the radioactive substance undergoes positron beta decay. The resulting positrons are further annihilated with electrons from the surrounding tissue, resulting in the emission of gamma-ray pairs, which are detected by the device, and then a 3D image is constructed based on the information obtained.
The selection of a radioactive isotope determines the biological process that one wishes to track in the research process. The process may be metabolism, transport of substances, etc. Behavior of the process, in turn, is the key to the correct diagnosis of the disease. The image above shows a tumor in the liver area.
But based on PET, it is difficult to understand in which part of the body the area with the maximum concentration of the radioactive substance is located. By combining body geometry (CT) and areas saturated with blood with a high concentration of radioactive substance (PET), we get:

Radioactive isotopes with different half-lives are used as a radioactive substance for PET. Fluorine-18 (fluorodeoxyglucose) is used to form all kinds of malignant tumors, iodine-124 is used to diagnose thyroid cancer, and gallium-68 is used to detect neuroendocrine tumors.
The Fusion functionality forms a new series in which the images of both modalities (both PET and CT) are combined. In the implementation, the images of both modalities are mixed, and then sorted along the Z axis (we assume that X and Y are the image axes). In fact, it turns out that the images in the series alternate (PET, CT, PET, CT ...). This series is further used to draw 2D fusion and 3D fusion. In the case of 2D fusion, images are drawn in pairs (PET-CT) in ascending order of Z:

In this case, the CT image was first drawn, then the PET.
3D fusion is implemented for a video card on CUDA. Both 3D-models - PET and CT are drawn on the video card at the same time and a real multimodal fusion is obtained. On the processor, the fusion also works, but it works a little differently. The fact is that on the processor both models are represented in memory as separate octo-trees. Therefore, when drawing it is necessary to trace two trees and synchronize the skipping of transparent voxels. And this would significantly reduce the speed of work. Therefore, it was decided to simply overlay the result of rendering one 3D-model on top of another.
4D CardiacCT
Cardiac CT technology is used to diagnose various disorders of the heart, including coronary heart disease, pulmonary thromboembolism and other diseases.
4D Cardiac CT is a 3D in time. Those. It turns out a small video, which we will call a film loop, in which each frame will be a 3D object. The source data is a set of dicom-images for all frames of a film loop at once. In order to convert a set of images into a film loop, you must first group the source images into frames, and then create 3D for each frame. Building a 3D object at the frame level is the same as for any series of dicom images. We use heuristic image sorting to group by frame, using the position of the image on the Z axis (assuming that X and Y are image axes). We believe that after grouping by frames, the same number of images is obtained in each frame. Switching the frame actually comes down to switching the 3D model.
5D Fusion Pet - CardiacCT
5D Fusion Pet - CardiacCT is a 4D Cardiac CT with the addition of fusion with PET as the fifth dimension. In the implementation, we first create two film loops: with CardiacCT and with PET. Then we make fuision of the corresponding frames of the movie band, which gives us a separate series. Then we build the resulting 3D series. It looks like this:
Virtual endoscopy
As an example of virtual endoscopy, we will consider virtual colonoscopy, since it is the most common type of virtual endoscopy. Virtual colonoscopy allows to construct a volumetric reconstruction of the abdominal cavity area on the basis of CT data and to make a diagnosis using this 3D reconstruction. In the viewer there is a fly-through camera tool with MPR navigation:

which also allows you to automatically follow the anatomical structure. In particular, it allows you to view the intraintestinal region automatically. Here's what it looks like:
The flight of the camera represents a series of consecutive movements in the intraintestinal region. For each step, the vector of camera movement to the next part of the anatomical structure is calculated. The calculation is based on transparent voxels in the next part of the anatomical structure. In fact, a certain voxel is calculated among transparent ones. The initial displacement vector is defined by the camera vector. In the Camera Flight tool, an exclusively perspective projection is used
There is also a functional for automatic intestinal segmentation, i.e. functionality to separate the intestinal region from the rest of the anatomy:

It is also possible to navigate through a segmented 3D model (the Show camera orientation button), which, by a mouse click on the 3D model, moves the camera to the appropriate position in the original anatomy.
Segmentation is implemented using a wave algorithm . It is believed that the anatomy is closed in the sense that it is not in contact with other organs and external space.
ECG Viewer (Waveform)
A separate module in the viewer implements data reading from the Waveform and its drawing. DICOM ECG Waveform is a special format for storing data from electrocardiogram leads defined by the DICOM standard. These electrocardiograms are twelve leads - 3 standard, 3 reinforced and 6 chest. The data of each lead is a sequence of measurements of the electrical voltage on the surface of the body. In order to draw the voltage, you need to know the scale vertically in mm / mV and the horizontal scale in mm / s:

As auxiliary attributes, the grid is also drawn for ease of measuring distances and a scale in the upper left corner. Scale options are selected taking into account medical practice: 10 and 20 mm / mV vertically, 25 and 50 mm / sec horizontally. Also implemented tools for measuring the distance horizontally and vertically.
DICOM-Viewer as a DICOM client
DICOM-Viewer, among other things, is a full-fledged DICOM client. It is possible to search on the PACS server, retrieve data from it, etc. The functions of the DICOM client are implemented using the open DCMTK library. Consider a typical use-case of the DICOM client on the example of the viewer. Perform a search for stages on a remote PACS server:

When a stage is selected, the series for the selected stage and the number of images in them are displayed at the bottom. The PACS server on which the search will be performed is indicated on the top right. The search can be parameterized by refining the search criteria: PID, study date, patient name, etc. The client search is performed by the C-FIND SCU command using the DCMTK library, which operates at one of the levels: STUDY, SERIES and IMAGE.
Next, images of the selected series can be downloaded using the C-GET-SCU and C-MOVE-SCU commands. The DICOM protocol obliges the parties to the connection, i.e. client and server, agree in advance what type of data they are going to transfer through this connection. A data type is a combination of the values of the SOPClassUID and TransferSyntax parameters. SOPClassUID determines the type of operation that is scheduled to be performed through this connection. The most commonly used SOPClassUIDs are: Verification SOP Class (ping server), Storage Service Class (image saving), Printer Sop Class (printing on a DICOM printer), CT Image Storage (saving CT images), MR Image Storage (saving image MRI) and others. TransferSyntax determines the format of a binary file. Popular TransferSyntaxs: Little Endian Explicit, Big Endian Implicit, JPEG Lossless Nonhierarchical (Processes 14). That is, in order to transfer MRI images in the Little Endian Implicit format, then MR Image Storage - Little Endian Explicit must be added to the connection.

Downloaded images are saved to the local storage and, when re-viewed, are downloaded from it, which allows to increase the performance of the viewer. Saved series are marked with a yellow icon in the upper left corner of the first image of the series.
Also, DicomViewer as a DICOM client can burn discs with studies in the DICOMDIR format. The DICOMDIR format is implemented as a binary file that contains relative paths to all DICOM files that are written to disk. Implemented using the DCMTK library. When reading a disc, the paths to all files from DICOMDIR are read and then loaded. To add stages and series to DICOMDIR, the following interface was developed:

That's all that I wanted to tell you about the functionality of DicomViewer. As always, feedback from qualified professionals is very welcome.
Viewer Link:
DICOM Viewer x86
DICOM Viewer x64
Data examples:
MANIX - for common examples (MPR, 2D, 3D, etc.)
COLONIX - for virtual colonoscopy
FIVIX - 4D CARDIAC-CT
CEREBRIX - Fusion PET-CT
Source: https://habr.com/ru/post/258621/
All Articles