Vectorization Advisor, another example - overclocking a fractal
We recently wrote about the new Vectorization Advisor. About what it is and why you need it, read the first article . The same post is devoted to the analysis of a specific example of optimizing an application using this tool.
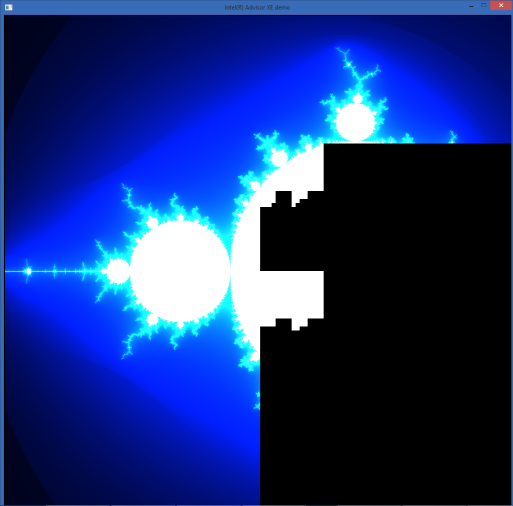
The application is taken from the examples of the Intel Threading Building Blocks (Intel TBB) library. It draws a Mandelbrot fractal and is parallelized in streams using Intel TBB. Those. it uses the advantages of a multi-core processor - let's see how things are with vector instructions.
Test system:
• Laptop with Intel Core i5-4300U (Haswell, 2 cores, 4 hardware streams)
• IDE: Microsoft Visual Studio 2013
• Compiler: Intel Composer 2015 update 2
• Intel Advisor XE 2016 Beta
• Appendix: Mandelbrot fractal, slightly modified. See <tbb_install_dir> \ examples \ task_priority
')
So, the first step: measure the basic version, running time: 4.51 seconds. Next, we launch a Survey analysis in Intel Advisor XE:
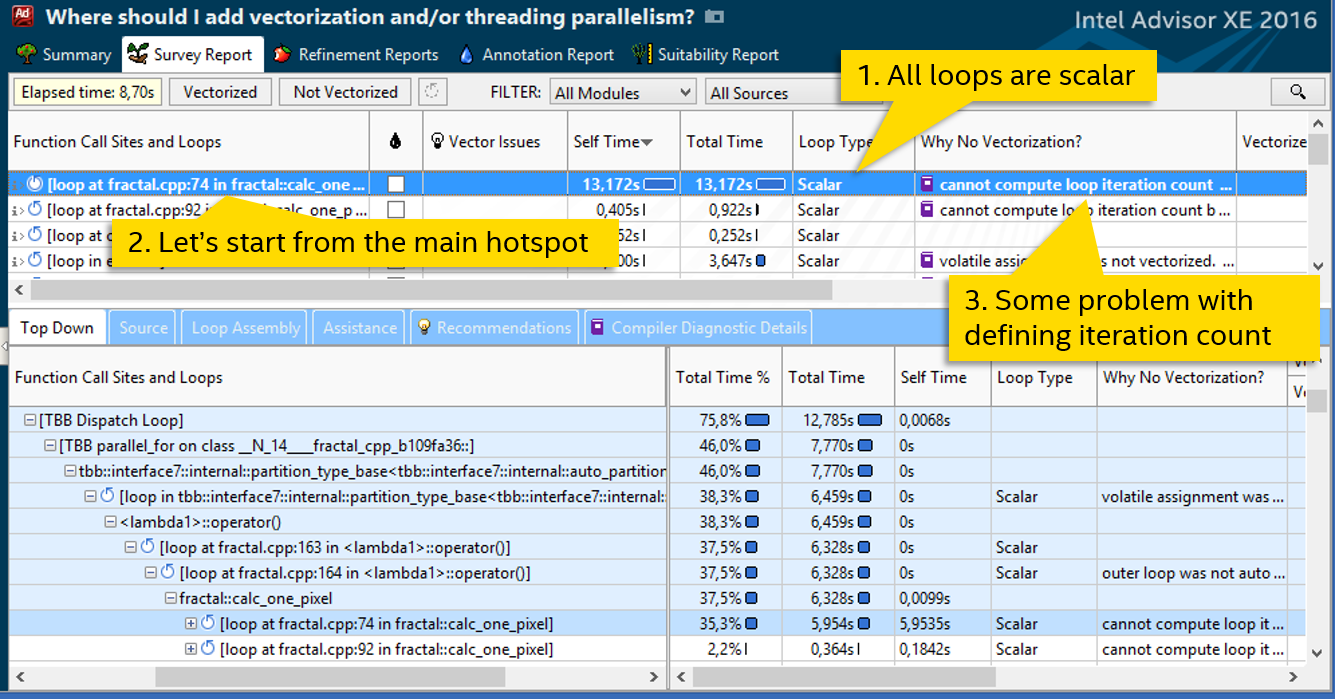
The column “Loop Type” says that all cycles are scalar, i.e. Do not use SIMD instructions. The most expensive cycle in the file fractal.cpp on line 74. It spends more than 13 seconds of processor time - and we’ll do it. The column “Why No Vectorization” contains a message stating that the compiler was unable to estimate the number of iterations of the cycle, and therefore did not vectorize it. Double click on the highlighted cycle, look at the code:
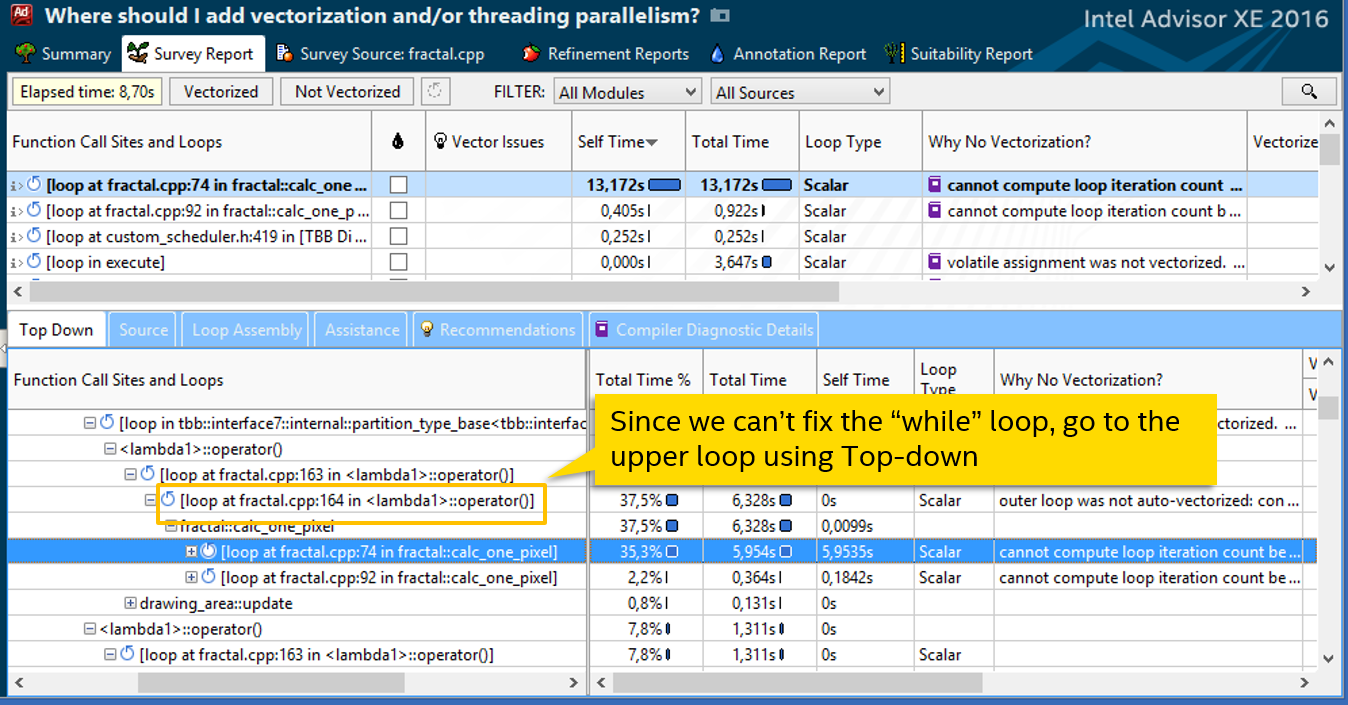
It becomes clear why the number of iterations is not known at compile time. Before diving into the details of the algorithm and trying to rewrite while, we will try to go in a simpler way. Let's see where our cycle comes from - the Top Down tab:
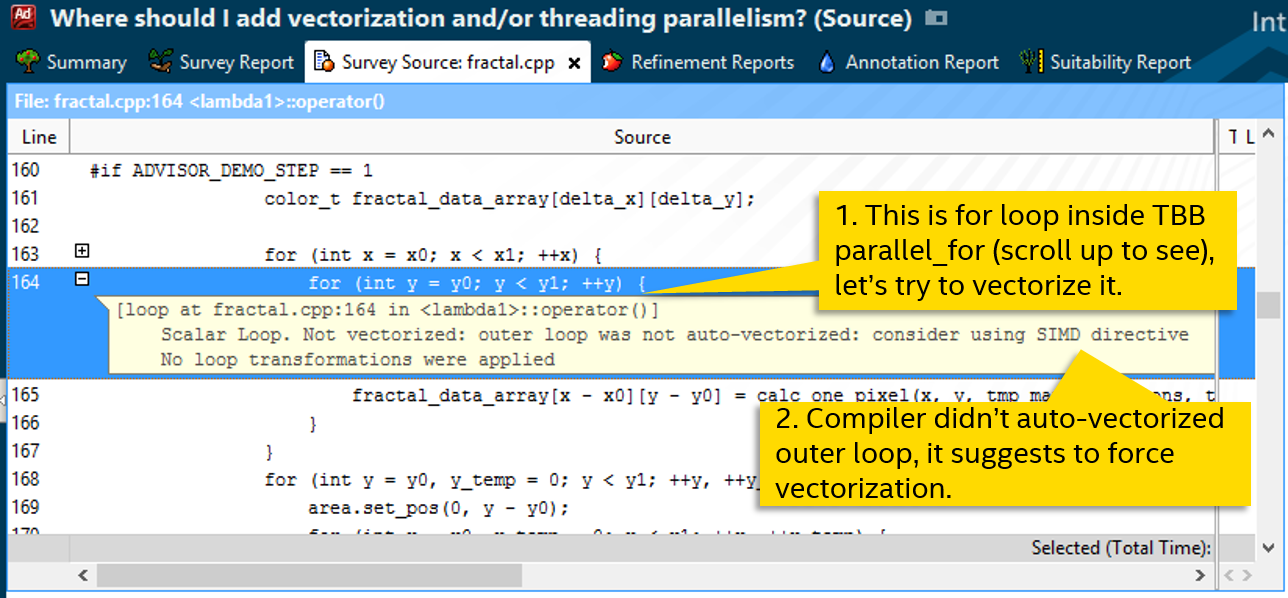
The next call cycle on line 164. This is a normal-looking for, which the compiler did not vectorize either (Scalar in the Loop Type column), probably not considering it effective. The diagnostic message suggests trying to force vectoring, for example, using the SIMD directive:
Follow the advice:
Reassemble the program and run Survey again. With #pragma simd, the cycle became not “Scalar”, but “Remainder”:

SIMD cycles, as you know, can be divided into peel, body and remainder. Body - in fact, a vectorized part, where several iterations are performed at once for a single instruction. Peel appears if the data is not aligned with the length of the vector, remainder - non-multiple iterations remaining at the end.
We only have Remainder. This means that the cycle was vectorized, but the body was not executed. The bookmark with recommendations speaks about the inefficiency of such a situation - it is necessary that more iterations be executed in the body, since the remainder here is a regular sequential cycle. Check how many iterations there are using Trip Counts analysis:

In our cycle, there are only 8 iterations, which is quite a bit. If there were more, a vectorized body would have something to perform. The line number has changed after code modifications, now it is line 179. Let's see how the number of iterations is determined:
The cycle of interest to us is called from the parallel loop of the Intel Threading Building Blocks library (Intel TBB). The iterations of the outer loop are distributed between different threads, each of which receives an object of type tbb :: blocked_range2d - its own local space of iterations. How small an iteration number can be in this space depends on the parameter, inner_grain_size . Those. The value of r.rows (). end () , which determines the number of loop iterations per line 179, depends on inner_grain_size .
We find in the code this same grain size (there are two for two nested parallel loops):
We are trying to increase the inner_grain_size to 32. Nothing bad is expected from this, just the division of work into Intel TBB threads will be more coarse-grained. See the result:
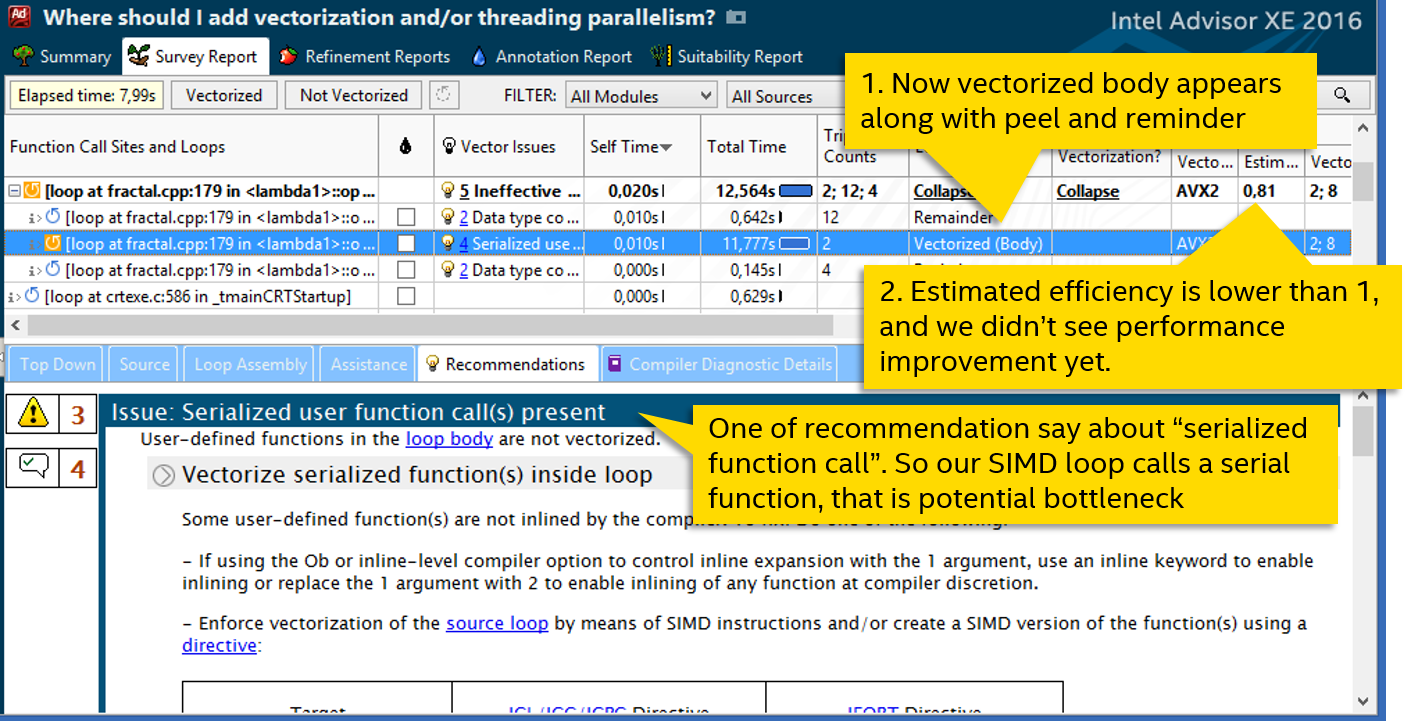
Now the vectorized body appears in the cycle, we finally achieved real use of SIMD instructions, the cycle is vectorized. But it's too early to rejoice - the performance has not grown at all.
We look at the recommendations of Advisor XE - one of them talks about calling a serialized (sequential) function. The fact is that if a vector cycle calls a function, then it needs a vectorized version that can accept parameters by vectors, and not by ordinary scalar variables. If the compiler could not generate such a vector function, then the regular, sequential version is used. And it is also called sequentially, nullifying the whole vectorization.
Look again at the loop code:
Fortunately, the function call here is one: calc_one_pixel . Since the compiler could not generate a vector version, we will try to help it. But first, let's look at the memory access patterns, this is useful for explicit vectoring:
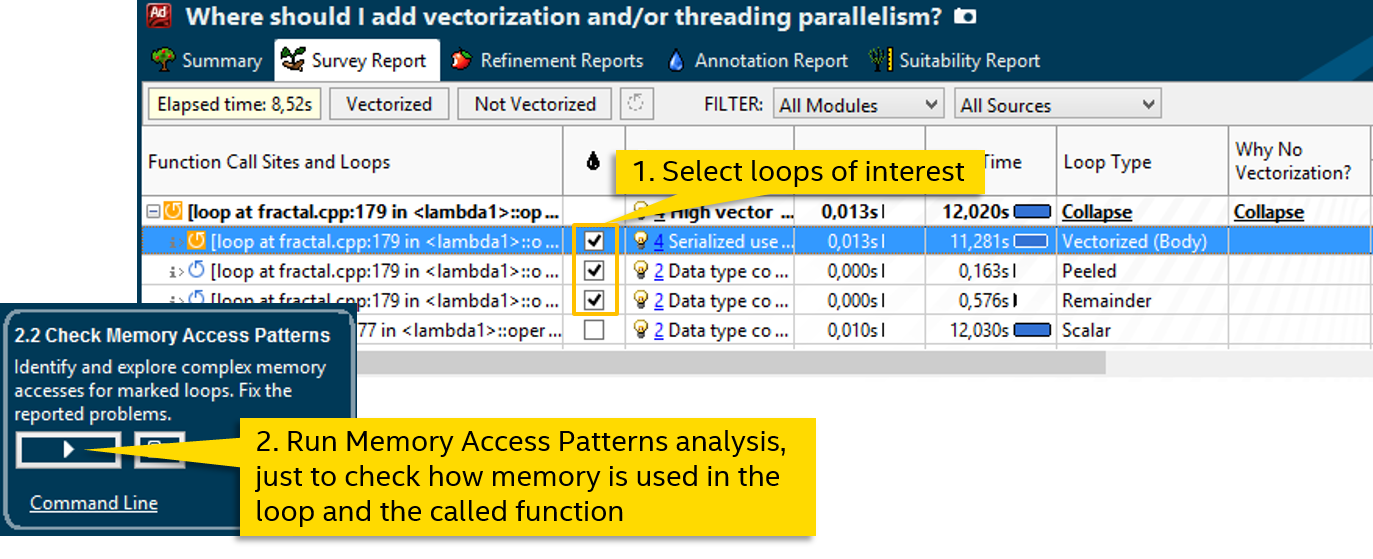
The analysis of the Memory Access Patterns of our cycle (along with the called function) tells us that all read or write operations are unit stride with a step of 0. That is, when accessing external data, the same variable is read or written from each iteration:
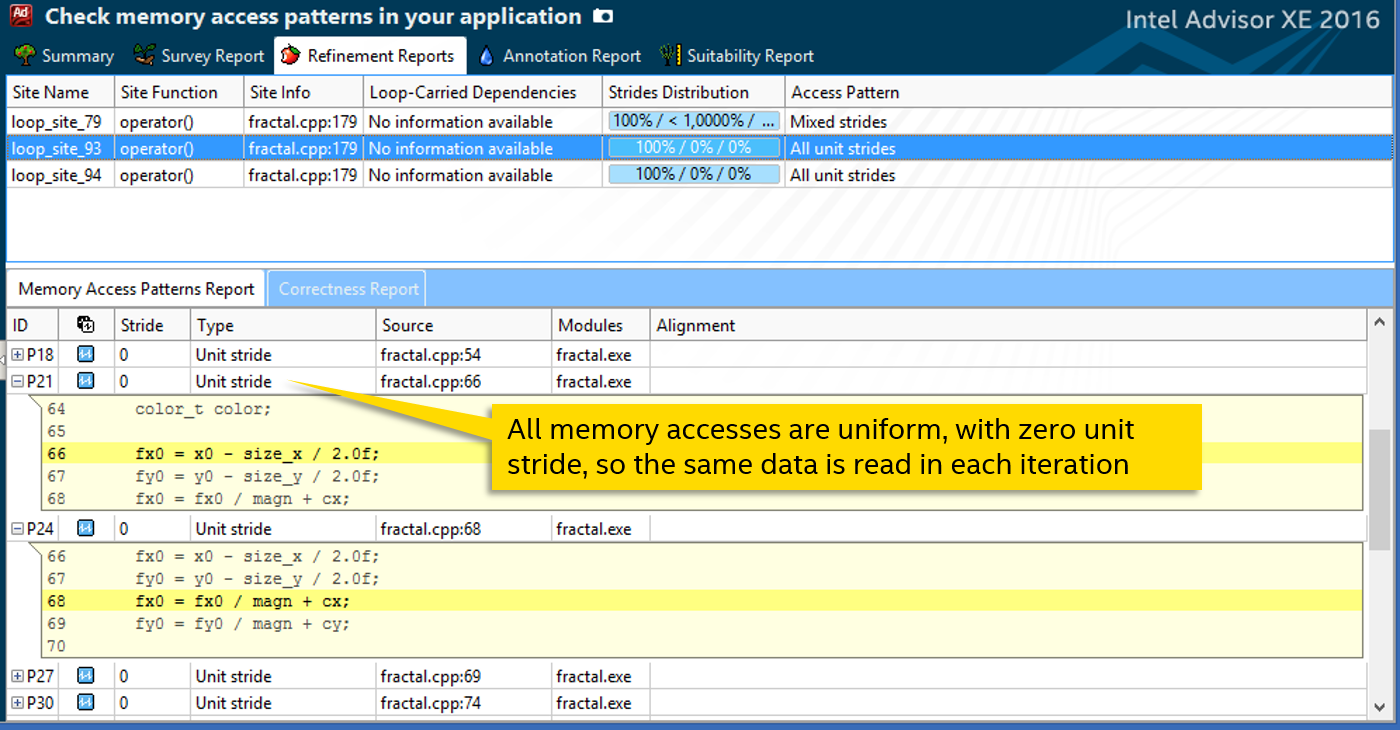
This knowledge is useful for us to manually vectorize the function. We will use the #pragma omp simd directive from the OpenMP 4 standard. It has options defining the parameter access pattern. For example, “linear” is used for monotonously growing quantities, mainly iterative variables. We are also suitable uniform - to access the same data.
By adding a directive on the definition of the function, we gave the compiler the opportunity to generate its vector version, which can process data arrays instead of the scalar declared. We get a noticeable increase in speed - the cycle is executed 7.5 seconds instead of 12:
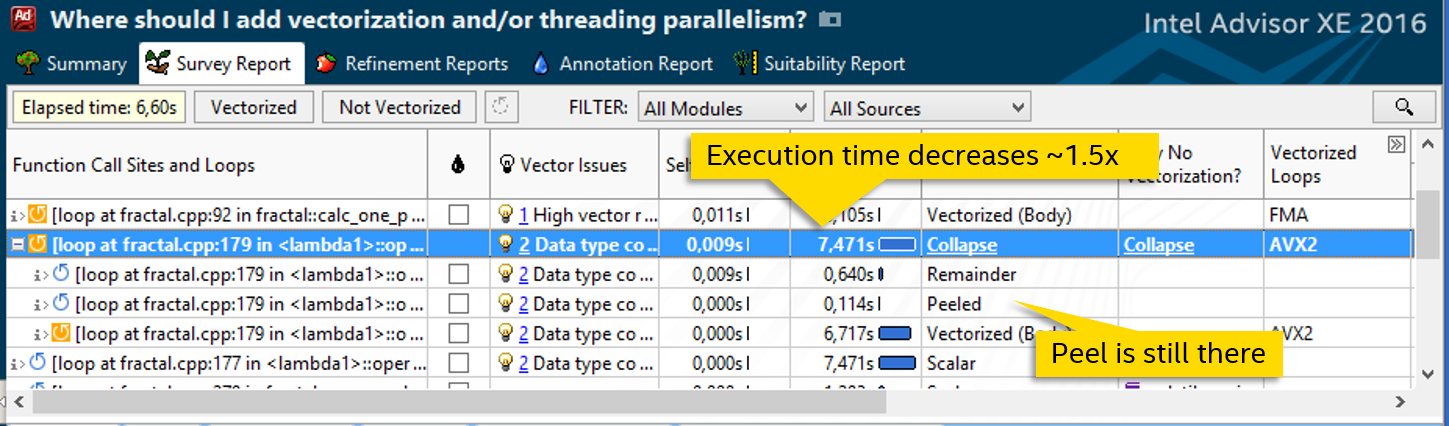
Let's try to fight other causes of the inefficiency of the vector cycle. The presence of a peel indicates that the data is not aligned. Add __declspec (align (32)) before defining an array in which the calc_one_pixel results are written :
After that peel disappears:

In the Advisor XE diagnostics table, you can pay attention to one more thing - the “Transformations” column says that the compiler has unrolled a cycle (unrolled) with a factor of 2:

Those. in order to optimize the number of iterations reduced by half. This in itself is not bad, but in the third step we specifically tried to increase them, and it turns out that the compiler does the opposite. Let's try to disable the scan with #pragma nounroll :
After this, the number of iterations is expected to double:
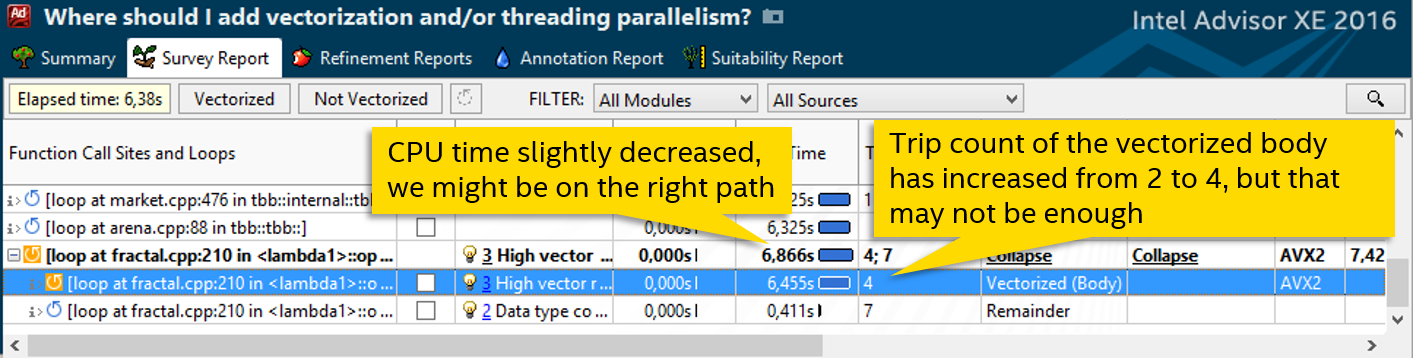
Manipulations with unrolling allowed to increase the number of iterations, but there was no improvement in performance. Let's see what happens if we increase grain_size even more, as in step 3. Just empirically test the optimal value:
It turns out to squeeze a little more, albeit quite a bit:

After all the manipulations, the test application runtime was reduced from 4.51 to 1.92 seconds, i.e. we achieved an acceleration of about 2.3 times. Let me remind you that our computational test has already been partially optimized - well parallelized by threads on Intel TBB, achieving good acceleration and scalability on multi-core processors. But due to the fact that the vectorization was not fully used, we still did not receive half of the possible performance. The first conclusion - vectorization can be of great importance , do not neglect it.
Now let's see the effect of various changes:

Forcing vectorization and increasing the number of iterations (steps 2 and 3) did not accelerate the program itself. We received the most important speed increase in step 4 after vectoring the function. However, this is the cumulative effect of steps 2-4. For a real cycle vectorization, it was necessary to increase the number of iterations, and force vectorization, and vectorize the function.
And the following steps had no special effect, in our case they could be completely skipped. Step 7 can be more or less attributed to success, and this is due to the fact that in step 3 we did not immediately set a greater number of iterations. However, the purpose of the post was to show the capabilities of Advisor XE with a specific example, so all the actions performed are described.
The application is taken from the examples of the Intel Threading Building Blocks (Intel TBB) library. It draws a Mandelbrot fractal and is parallelized in streams using Intel TBB. Those. it uses the advantages of a multi-core processor - let's see how things are with vector instructions.

Test system:
• Laptop with Intel Core i5-4300U (Haswell, 2 cores, 4 hardware streams)
• IDE: Microsoft Visual Studio 2013
• Compiler: Intel Composer 2015 update 2
• Intel Advisor XE 2016 Beta
• Appendix: Mandelbrot fractal, slightly modified. See <tbb_install_dir> \ examples \ task_priority
')
1. Assess the situation
So, the first step: measure the basic version, running time: 4.51 seconds. Next, we launch a Survey analysis in Intel Advisor XE:

The column “Loop Type” says that all cycles are scalar, i.e. Do not use SIMD instructions. The most expensive cycle in the file fractal.cpp on line 74. It spends more than 13 seconds of processor time - and we’ll do it. The column “Why No Vectorization” contains a message stating that the compiler was unable to estimate the number of iterations of the cycle, and therefore did not vectorize it. Double click on the highlighted cycle, look at the code:
while ( ((x*x + y*y) <= 4) && (iter < max_iterations) ) { xtemp = x*x - y*y + fx0; y = 2 * x*y + fy0; x = xtemp; iter++; } It becomes clear why the number of iterations is not known at compile time. Before diving into the details of the algorithm and trying to rewrite while, we will try to go in a simpler way. Let's see where our cycle comes from - the Top Down tab:

2. We force vectorization
The next call cycle on line 164. This is a normal-looking for, which the compiler did not vectorize either (Scalar in the Loop Type column), probably not considering it effective. The diagnostic message suggests trying to force vectoring, for example, using the SIMD directive:

Follow the advice:
#pragma simd // forced vectorization for (int y = y0; y < y1; ++y) { fractal_data_array[x - x0][y - y0] = calc_one_pixel(x, y, tmp_max_iterations, tmp_size_x, tmp_size_y, tmp_magn, tmp_cx, tmp_cy, 255); } Reassemble the program and run Survey again. With #pragma simd, the cycle became not “Scalar”, but “Remainder”:

SIMD cycles, as you know, can be divided into peel, body and remainder. Body - in fact, a vectorized part, where several iterations are performed at once for a single instruction. Peel appears if the data is not aligned with the length of the vector, remainder - non-multiple iterations remaining at the end.
We only have Remainder. This means that the cycle was vectorized, but the body was not executed. The bookmark with recommendations speaks about the inefficiency of such a situation - it is necessary that more iterations be executed in the body, since the remainder here is a regular sequential cycle. Check how many iterations there are using Trip Counts analysis:

3. Increasing the number of iterations
In our cycle, there are only 8 iterations, which is quite a bit. If there were more, a vectorized body would have something to perform. The line number has changed after code modifications, now it is line 179. Let's see how the number of iterations is determined:
tbb::parallel_for(tbb::blocked_range2d<int>(y0, y1, inner_grain_size, x0, x1, inner_grain_size), [&] (tbb::blocked_range2d<int> &r){ int x0 = r.cols().begin(), y0 = r.rows().begin(), x1 = r.cols().end(), y1 = r.rows().end(); ... for (int x = x0; x < x1; ++x) { for (int y = y0; y < y1; ++y) { // 179 fractal_data_array[x - x0][y - y0] = calc_one_pixel(x, y, tmp_max_iterations, tmp_size_x, tmp_size_y, tmp_magn, tmp_cx, tmp_cy, 255); } } ... The cycle of interest to us is called from the parallel loop of the Intel Threading Building Blocks library (Intel TBB). The iterations of the outer loop are distributed between different threads, each of which receives an object of type tbb :: blocked_range2d - its own local space of iterations. How small an iteration number can be in this space depends on the parameter, inner_grain_size . Those. The value of r.rows (). end () , which determines the number of loop iterations per line 179, depends on inner_grain_size .
We find in the code this same grain size (there are two for two nested parallel loops):
int grain_size = 64; int inner_grain_size = 8; We are trying to increase the inner_grain_size to 32. Nothing bad is expected from this, just the division of work into Intel TBB threads will be more coarse-grained. See the result:

Now the vectorized body appears in the cycle, we finally achieved real use of SIMD instructions, the cycle is vectorized. But it's too early to rejoice - the performance has not grown at all.
4. Vectorize the function
We look at the recommendations of Advisor XE - one of them talks about calling a serialized (sequential) function. The fact is that if a vector cycle calls a function, then it needs a vectorized version that can accept parameters by vectors, and not by ordinary scalar variables. If the compiler could not generate such a vector function, then the regular, sequential version is used. And it is also called sequentially, nullifying the whole vectorization.
Look again at the loop code:
for (int y = y0; y < y1; ++y) { // 179 fractal_data_array[x - x0][y - y0] = calc_one_pixel(x, y, tmp_max_iterations, tmp_size_x, tmp_size_y, tmp_magn, tmp_cx, tmp_cy, 255); } Fortunately, the function call here is one: calc_one_pixel . Since the compiler could not generate a vector version, we will try to help it. But first, let's look at the memory access patterns, this is useful for explicit vectoring:

The analysis of the Memory Access Patterns of our cycle (along with the called function) tells us that all read or write operations are unit stride with a step of 0. That is, when accessing external data, the same variable is read or written from each iteration:

This knowledge is useful for us to manually vectorize the function. We will use the #pragma omp simd directive from the OpenMP 4 standard. It has options defining the parameter access pattern. For example, “linear” is used for monotonously growing quantities, mainly iterative variables. We are also suitable uniform - to access the same data.
#pragma omp declare simd uniform(x0, max_iterations, size_x, size_y, magn, cx, cy, gpu) color_t fractal::calc_one_pixel(float x0, float y0, int max_iterations, int size_x, int size_y, float magn, float cx, By adding a directive on the definition of the function, we gave the compiler the opportunity to generate its vector version, which can process data arrays instead of the scalar declared. We get a noticeable increase in speed - the cycle is executed 7.5 seconds instead of 12:

5. Align the data
Let's try to fight other causes of the inefficiency of the vector cycle. The presence of a peel indicates that the data is not aligned. Add __declspec (align (32)) before defining an array in which the calc_one_pixel results are written :
__declspec(align(32)) color_t fractal_data_array[delta_x][(delta_y / 16 + 1) * 16]; // aligned data for (int x = x0; x < x1; ++x) { #pragma simd for (int y = y0; y < y1; ++y) { fractal_data_array[x - x0][y - y0] = calc_one_pixel(x, y, tmp_max_iterations, tmp_size_x, tmp_size_y, tmp_magn, tmp_cx, tmp_cy, 255); } } After that peel disappears:

6. Remove the unrolling cycle
In the Advisor XE diagnostics table, you can pay attention to one more thing - the “Transformations” column says that the compiler has unrolled a cycle (unrolled) with a factor of 2:

Those. in order to optimize the number of iterations reduced by half. This in itself is not bad, but in the third step we specifically tried to increase them, and it turns out that the compiler does the opposite. Let's try to disable the scan with #pragma nounroll :
#pragma simd #pragma nounroll // added unrolling for (int y = y0; y < y1; ++y) { fractal_data_array[x - x0][y - y0] = calc_one_pixel(x, y, tmp_max_iterations, tmp_size_x, tmp_size_y, tmp_magn, tmp_cx, tmp_cy, 255); } After this, the number of iterations is expected to double:

7. Increasing the number of iterations even more.
Manipulations with unrolling allowed to increase the number of iterations, but there was no improvement in performance. Let's see what happens if we increase grain_size even more, as in step 3. Just empirically test the optimal value:
int grain_size = 256; // increase from 64 int inner_grain_size = 64; // increase from 8 It turns out to squeeze a little more, albeit quite a bit:

8. Results
After all the manipulations, the test application runtime was reduced from 4.51 to 1.92 seconds, i.e. we achieved an acceleration of about 2.3 times. Let me remind you that our computational test has already been partially optimized - well parallelized by threads on Intel TBB, achieving good acceleration and scalability on multi-core processors. But due to the fact that the vectorization was not fully used, we still did not receive half of the possible performance. The first conclusion - vectorization can be of great importance , do not neglect it.
Now let's see the effect of various changes:

Forcing vectorization and increasing the number of iterations (steps 2 and 3) did not accelerate the program itself. We received the most important speed increase in step 4 after vectoring the function. However, this is the cumulative effect of steps 2-4. For a real cycle vectorization, it was necessary to increase the number of iterations, and force vectorization, and vectorize the function.
And the following steps had no special effect, in our case they could be completely skipped. Step 7 can be more or less attributed to success, and this is due to the fact that in step 3 we did not immediately set a greater number of iterations. However, the purpose of the post was to show the capabilities of Advisor XE with a specific example, so all the actions performed are described.
Source: https://habr.com/ru/post/257309/
All Articles