Hybrid implementation of the MST algorithm using CPU and GPU
Introduction
Solving the problem of finding minimum spanning trees (MST - minimum spanning tree) is a common task in various areas of research: recognition of various objects, computer vision, analysis and construction of networks (for example, telephone, electrical, computer, road, etc.), chemistry and biology and many others. There are at least three known algorithms that solve this problem: Boruvka, Kruskal, and Prima. The processing of large graphs (occupying several GB) is quite a laborious task for the central processing unit (CPU) and is currently in demand. Graphic accelerators (GPUs), which are capable of showing much higher performance than CPU, are becoming more widespread. But the MST task, like many graph processing tasks, doesn’t fall well on the GPU architecture. This article will discuss the implementation of this algorithm on the GPU. It will also show how the CPU can be used to build a hybrid implementation of this algorithm on the shared memory of one node (consisting of a GPU and several CPUs).
Description of the graph representation format
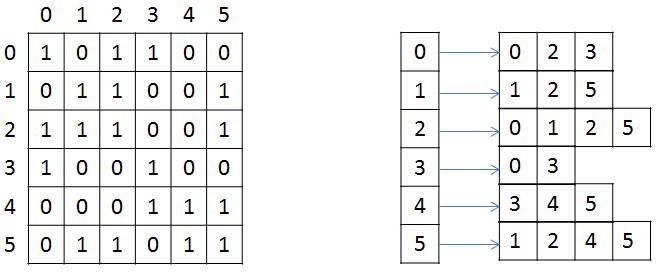
We briefly consider the storage structure of an undirected weighted graph, since in the future it will be mentioned and transformed. The graph is set in compressed CSR (Compressed Sparse Row) [1] format. This format is widely used to store sparse matrices and graphs. For a graph with N vertices and M edges, three arrays are needed: X, A, and W. The array X is of size N + 1, the other two are 2 * M, since in an undirected graph, for any pair of vertices, it is necessary to store the forward and reverse arcs. The array X stores the beginning and end of the list of neighbors that are stored in array A, that is, the entire list of neighbors of J is in array A from index X [J] to X [J + 1], not including it. Similar indexes store the weights of each edge from the vertex J. For illustration, the graph below shows a graph of 6 vertices, recorded using an adjacency matrix, and on the right, in CSR format (for simplicity, the weight of each edge is not specified).
Test graphs
Immediately I will describe on which graphs the testing took place, since the description of the transformation algorithms and the MST algorithm will require knowledge of the structure of the graphs in question. Two types of synthetic graphs are used to evaluate implementation performance: RMAT-graphs and SSCA2-graphs. R-MAT-graphs well simulate real graphs from social networks, the Internet [2] . In this case, RMAT-graphs are considered with an average degree of connectivity of a vertex 32, and the number of vertices is a power of two. In such an RMAT-graph there is one large connected component and a number of small connected components or hanging vertices. SSCA2-graph is a large set of independent components, connected by edges with each other [3] . The SSCA2-graph is generated so that the average degree of connectivity of a vertex is close to 32, and its number of vertices is also a power of two. Thus, two completely different in structure of the graph are considered.
')
Input Conversion
Since the algorithm will be tested on the RMAT and SSCA2 columns, which are obtained using a generator, some transformations need to be done to improve the algorithm performance. All conversions will not be taken into account in the performance calculation.
- Local Sorting Vertex List
For each vertex, we sort its list of neighbors by weight, in ascending order. This will partially simplify the selection of the minimum edge at each iteration of the algorithm. Since this sorting is local, it does not provide a complete solution to the problem. - Renumbering all vertices of the graph
Enumerate the vertices of the graph in such a way that the most connected vertices have the closest numbers. As a result of this operation, in each connected component, the difference between the maximum and minimum vertex numbers will be the smallest, which will make the best use of the small cache of the graphic process. It should be noted that for RMAT graphs, this renumbering does not have a significant effect, since in this graph there is a very large component that does not fit in the cache even after applying this optimization. For SSCA2 graphs, the effect of this transformation is more noticeable, since in this graph there are a large number of small components. - Mapping graph weights to integers
In this problem, we do not need to perform any operations on the weights of the graph. We need to be able to compare the weights of two ribs. For these purposes, you can use integers, instead of double precision numbers, since the processing speed of single precision numbers on the GPU is much higher than double. This conversion can be performed for graphs in which the number of unique edges does not exceed 2 ^ 32 (the maximum number of different numbers that fit in an unsigned int). If the average degree of connectivity of each vertex is 32 m, then the largest graph that can be processed using this transformation will have 2 ^ 28 vertices and will occupy 64 GB in memory. To date, the largest amount of memory in the NVidia Tesla k40 [4] / NVidia Titan X [5] and AMD FirePro w9100 [6] accelerators is 12GB and 16GB, respectively. Therefore, on one GPU using this conversion, it is possible to process rather large graphs. - Compressing vertex information
This transformation applies only to SSCA2 graphs due to their structure. In this task, memory performance of all levels plays a decisive role: from global memory to the first-level cache. To reduce the traffic between the global memory and L2 cache, you can store information about the vertices in a compressed form. Initially, information about vertices is represented as two arrays: array X, which stores the beginning and end of the list of neighbors in array A (an example of only one vertex):
J has 10 vertices-neighbors, and if the number of each neighbor is stored using the unsigned int type, then to store the list of J-neighbors, 10 * sizeof (unsigned int) bytes is required, and for the whole graph - 2 * M * sizeof (unsigned int) bytes. We assume that sizeof (unsigned int) = 4 bytes, sizeof (unsigned short) = 2 bytes, sizeof (unsigned char) = 1 byte. Then for this vertex you need 40 bytes to store the list of neighbors.
It is not difficult to notice that the difference between the maximum and minimum vertex numbers in this list is 8, and only 4 bits are needed to store this number. Based on the considerations that the difference between the maximum and minimum vertex numbers may be less than an unsigned int, you can imagine the number of each vertex as follows:
base_J + 256 * k + short_endV,
where base_J is, for example, the minimum vertex number from the entire list of neighbors. In this example, it will be 1. This variable will be of type unsigned int and there will be as many such variables as there are vertices in the graph; Next, we calculate the difference between the number of the vertex and the selected base. Since we chose the smallest peak as the base, this difference will always be positive. For the SSCA2 graph, this difference will be placed in an unsigned short. short_endV is the remainder of dividing by 256. To store this variable, we will use the unsigned char type; and k is the integer part of division by 256. For k, select 2 bits (that is, k lies in the range from 0 to 3). The selected representation is sufficient for the graph under consideration. In bit representation, it looks like this:
Thus, to store the list of vertices, (1 + 0.25) * 10 + 4 = 16.5 bytes is required for this example, instead of 40 bytes, and for the whole graph: (2 * M + 4 * N + 2 * M / 4 ) instead of 2 * M * 4. If N = 2 * M / 32, then the total volume will decrease in
(8 * M) / (2 * M + 8 * M / 32 + 2 * M / 4) = 2.9 times
General description of the algorithm
For the implementation of the MST algorithm Boruvka algorithm was chosen. The basic description of Boruvka's algorithm and the illustration of its iterations are well represented at this link [7] .
According to the algorithm, all vertices are initially included in the minimum tree. Next, you must perform the following steps:
- Find the minimum edges between all the trees for their subsequent union. If no edge is selected at this step, then the task response is received.
- Join the appropriate trees. This step is divided into two stages: deletion of cycles, since two trees can indicate each other as a candidate for a merge, and a merge stage when the number of a tree is selected that includes the subtrees to be joined. For definiteness, we will choose the minimum number. If only one tree remains in the course of the merge, the answer of the problem is received.
- Renumber the received trees to go to the first step (so that all trees have numbers from 0 to k)
Algorithm Stages
In general, the implemented algorithm is as follows:
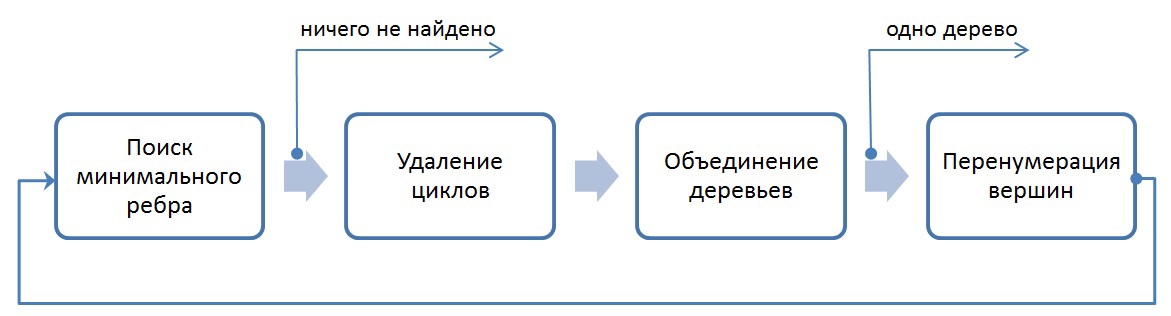
The exit from the entire algorithm occurs in two cases: if all vertices after N iterations are combined into one tree, or if it is impossible to find the minimum edge from each tree (in this case, the minimum spanning trees are found).
1. Search for the minimum edge.
First, each vertex of the graph is placed in a separate tree. Next comes the iterative process of merging trees, consisting of the four procedures discussed above. The procedure for finding the minimum edge allows you to select exactly those edges that will be included in the minimum spanning tree. As described above, at the input to this procedure, the converted graph is stored in CSR format. Since for the list of neighbors partial edges were sorted by weight, the choice of the minimum vertex is reduced to viewing the list of neighbors and selecting the first vertex that belongs to another tree. If we assume that there are no loops in the graph, then at the first step of the algorithm, choosing the minimum vertex reduces to selecting the first vertex from the list of neighbors for each vertex under consideration, because the list of neighboring vertices (which make up the edges of the graph together with the vertex under consideration) is sorted by weight edges and each vertex is included in a separate tree. At any other step, you need to view a list of all adjacent vertices in order and select the vertex that belongs to another tree.
Why is it impossible to choose the second vertex from the list of neighboring vertices and put this edge as minimal? After the procedure of merging trees (which will be discussed later), a situation may arise that some vertices from the neighboring list may end up in the same tree as the considered one, thus this edge will be a loop for this tree, and according to the condition of the algorithm it is necessary to choose the minimum edge to other trees.
Union Find [8] is well suited to implement vertex processing and perform the search, merge, and merge procedure. Unfortunately, not all structures are optimally processed on the GPU. The most beneficial in this task (as in most others) is to use continuous arrays in the memory of the GPU, instead of linked lists. Below we will consider similar algorithms for finding the minimum edge, combining segments, deleting cycles in a graph.
Consider the algorithm for finding the minimum edge. It can be presented in two steps:
- selection of the minimum edge outgoing from each vertex (which is included in some segment) of the graph under consideration;
- selection of the edge of the minimum weight for each tree.
In order not to move the vertex information recorded in CSR format, we will use two auxiliary arrays, which will store the index of the beginning and end of the A list of the neighbors list. Two array data will denote segments of lists of vertices belonging to the same tree. For example, in the first step, the array of beginnings or lower values will have the values 0..N of the array X, and the array of the ends or upper values will have the values 1..N + 1 of the array X. And then, after the procedure of merging trees (which will be discussed later ), these segments are mixed, but the array of neighbors A will not be changed in memory.
Both steps can be performed in parallel. To perform the first step, you need to view the list of neighbors of each vertex (or each segment) and select the first edge belonging to another tree. You can select one warp (consisting of 32 threads) to view the list of neighbors of each vertex. It should be remembered that several segments of the array of neighboring vertices A may not lie in a row and belong to the same tree (the segments belonging to tree 0 and the green to tree 1 are highlighted in red):
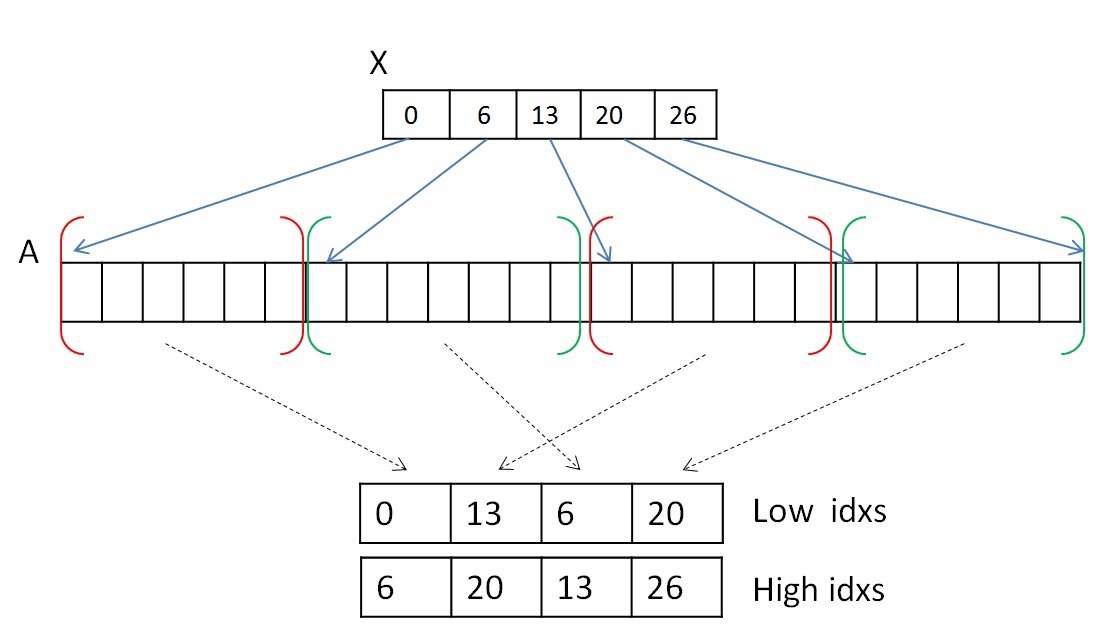
Since each segment of the list of neighbors is sorted, it is not necessary to look through all the vertices. Since one warp consists of 32 threads, it will be viewed in portions of 32 vertices. After the 32 vertices are viewed, the result must be combined, and if nothing is found, then view the next 32 vertices. To combine the result, you can use the scan algorithm [9] . This algorithm can be implemented within one warp using shared memory or using new shfl instructions [10] (available from the Kepler architecture), which allow you to exchange data between threads of a single warp for one instruction. As a result of the experiments, it turned out that shfl instructions allow speeding up the work of the entire algorithm approximately two times. Thus, this operation can be performed using shfl instructions, for example, like this:
unsigned idx = blockIdx.x * blockDim.x + threadIdx.x; // unsigned lidx = idx % 32; #pragma unroll for (int offset = 1; offset <= 16; offset *= 2) { tmpv = __shfl_up(val, (unsigned)offset); if(lidx >= offset) val += tmpv; } tmpv = __shfl(val, 31); // . 1, - // , . As a result of this step, the following information will be recorded for each segment: the number of the vertex in array A, which is included in the edge of the minimum weight and the weight of the edge itself. If nothing is found, then the number of the vertex can be written, for example, the number N + 2.
The second step is needed to reduce the selected information, namely, the choice of the edge with the minimum weight for each of the trees. This step is done due to the fact that the segments belonging to the same tree are viewed in parallel and independently, and an edge of minimum weight is selected for each of the segments. In this step, one warp can reduce the information for each tree (across several segments) and shfl instructions can also be used for the reduction. After completing this step, it will be known with which tree each of the trees is connected by a minimum edge (if it exists). To record this information, we introduce two more auxiliary arrays, in one of which we will store the numbers of the trees, to which there is a minimum edge, in the second - the number of the vertex in the source graph, which is the root of the vertices in the tree. The result of this step is illustrated below:
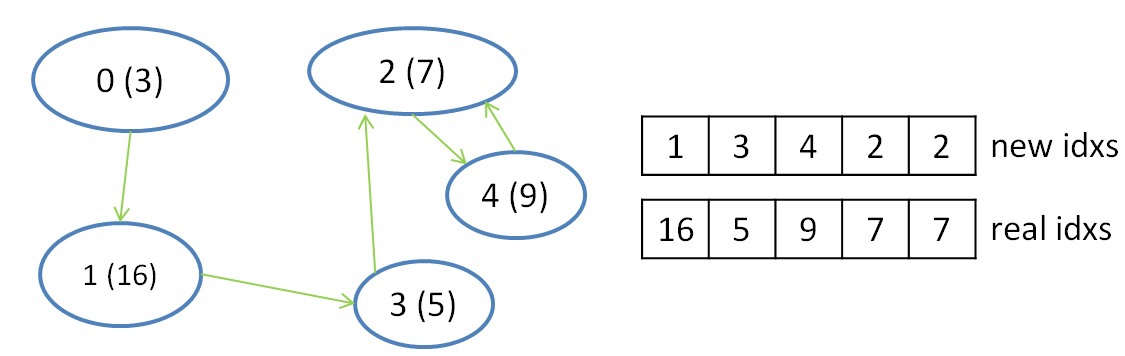
It should be noted that to work with indices, two more arrays are needed, which help to convert the initial indices into new indices and get the initial one using the new index. These so-called index conversion tables are updated with each iteration of the algorithm. The table for obtaining the new index according to the original index has the size N - the number of vertices in the graph, and the table for obtaining the initial index is reduced by each iteration and has a size equal to the number of trees at any selected iteration of the algorithm (at the first iteration of the algorithm this table also has size n).
2. Deleting loops.
This procedure is required to remove loops between two trees. This situation occurs when the tree N1 has the minimum edge to the tree N2, and the tree N2 has the minimum edge to the tree N1. In the picture above, there is a cycle only between two trees with numbers 2 and 4. Since there are fewer trees at each iteration, we will choose the minimum number from the two trees that make up the cycle. In this case, 2 will point to 2, and 4 will continue to point to 2. With such checks, you can define such a cycle and eliminate it in favor of the minimum number:
unsigned i = blockIdx.x * blockDim.x + threadIdx.x; unsigned local_f = F[i]; if (F[local_f] == i) { if (i < local_f) { F[i] = i; . . . . . . . } } This procedure can be performed in parallel, since each vertex can be processed independently and records to a new array of vertices without cycles do not overlap.
3. Combining trees.
This procedure combines the trees into larger ones. The procedure for deleting cycles between two trees is essentially a preprocessing before this procedure. It avoids looping when merging trees. Merging trees is the process of choosing a new root by changing links. If we assume tree 0 points to tree 1, and in turn tree 1 points to tree 3, then you can change the link of tree 0 from tree 1 to tree 3. This link change should be made if changing the link does not cause a cycle between two trees . Considering the example above, after the procedures for deleting cycles and merging trees, there will be only one tree number 2. The merging process can be represented like this:
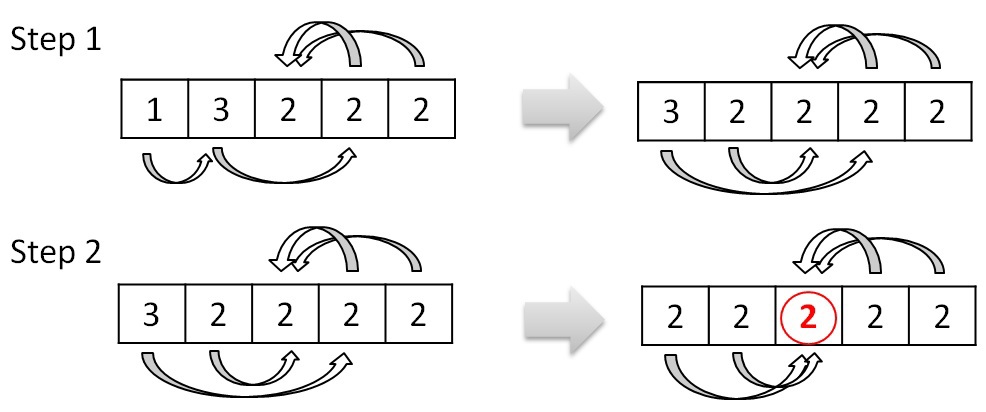
The structure of the graph and the principle of its processing is such that there will be no situation when the procedure will loop and it can also be executed in parallel.
4. Renumbering vertices (trees).
After the merge procedure is completed, it is necessary to renumber the obtained trees so that their numbers go in a row from 0 to P. By construction, the new numbers should receive array elements that satisfy the condition F [i] == i (for the above example, only the element with index 2). Thus, with the help of atomic operations, you can mark the entire array with new values from 1 ... (P + 1). Next, fill in the tables for obtaining a new index on the original and initial index on the new:
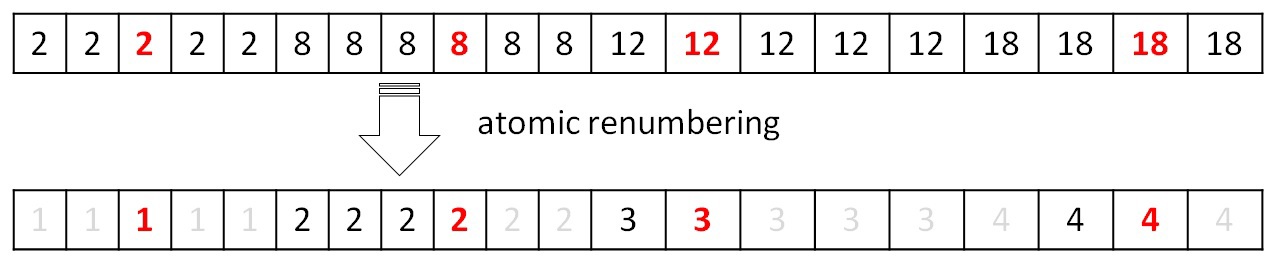
Working with these tables is described in the procedure for finding the minimum edge. The next iteration cannot be correctly performed without updating the table data. All described operations are performed in parallel and on the GPU.
Let's summarize a little . All 4 procedures are performed in parallel and on a graphics accelerator. Work is carried out with one-dimensional arrays. The only difficulty is that in all these procedures there is an indirect indexing. And in order to reduce cache misses from such work with arrays, various graph permutations were used, described at the very beginning. But, unfortunately, not for every graph can reduce losses from indirect indexation. As will be shown later, with this approach, not very high performance is achieved on RMAT-graphs. The search for the minimum edge takes up to 80% of the running time of the entire algorithm, while the rest accounts for the remaining 20%. This is due to the fact that in the procedures of merging, deletion of cycles and renumbering of vertices, work is carried out with arrays whose length is constantly decreasing (from iteration to iteration). For the considered graphs, it is necessary to do about 7-8 iterations. This means that the number of processed vertices already at the first step becomes much smaller than N / 2. While in the basic procedure for finding the minimum edge, we work with arrays of vertices A and an array of weights W (although certain elements are selected).
In addition to the storage of the graph, several more arrays of length N were used:
- array of lower values and array of upper values. Used to work with segments of array A;
- an array table to get the initial index in a new way;
- array table to get the new index on the original;
- array for numbers of vertices and an array for the corresponding weights, used in the second step of the procedure for finding the minimum edge;
- an auxiliary array that allows you to determine in the first step of the procedure for finding the minimum edge to which tree a particular segment belongs.
Hybrid implementation of the procedure for finding the minimum edge.
The algorithm described above ultimately is not badly executed on a single GPU. The solution to this problem is organized in such a way that you can try to parallelize this procedure also on the CPU. Of course, this can be done only on shared memory, and for this purpose I used the OpenMP standard and data transfer between the CPU and GPU over the PCIe bus. If we present the execution of procedures at one iteration on the time line, then the picture when using one GPU will be approximately like this:

Initially, all graph data is stored both on the CPU and on the GPU. In order for the CPU to read, it is necessary to transmit information about the segments that were moved during the merging of the trees. Also, in order for the GPU to continue the iteration of the algorithm, it is necessary to return the calculated data. It would be logical to use asynchronous copying between the host and the accelerator:

The algorithm on the CPU repeats the algorithm used on the GPU, only Openpa is used to parallelize the loop [11] . As one would expect, the CPU does not count as quickly as the GPU, and the overhead of copying also interferes. In order for the CPU to calculate its part, the data for calculation should be divided in the ratio of 1: 5, that is, no more than 20% -25% should be sent to the CPU, and the rest should be calculated to the GPU. The rest of the procedures are not profitable to count both there and there, since they take very little time, and the overhead and slow CPU speed only increase the algorithm time. Also very important is the speed of copying between the CPU and the GPU. On the tested platform, PCIe 3.0 was supported, which allowed to reach 12GB / s.
Today, the amount of RAM on the GPU and CPU differs significantly in favor of the latter. On the test platform, 6 GB GDDR5 was installed, while the CPU had 48 GB. Restrictions on memory on the GPU do not allow cheating large graphs. And here we can be helped by the CPU and the Unified Memory technology [12] , which allows us to handle the GPU in the CPU memory. Since the graph information is only needed in the search for the minimum edge, for large graphs, you can do the following: first place all auxiliary arrays in the GPU memory, and then place some of the graph arrays (the array of neighbors A, array X and the array of weights W) in memory GPU, but what does not fit - in the memory of the CPU. Further, during the counting, data can be divided so that the part that does not fit on the GPU is processed on the CPU, and the GPU minimally uses the access to the CPU memory (since the access to the CPU memory from the graphics accelerator is done via the PCIe bus at a speed of no more than 15 GB / s) It is known in advance in what proportion the data was divided, so in order to determine which memory you need to contact - in a GPU or CPU - you just need to enter a constant indicating the point at which the arrays are separated and with a single check in the algorithm on the GPU you can determine where to go appeal The location in the data memory of arrays can be represented like this:
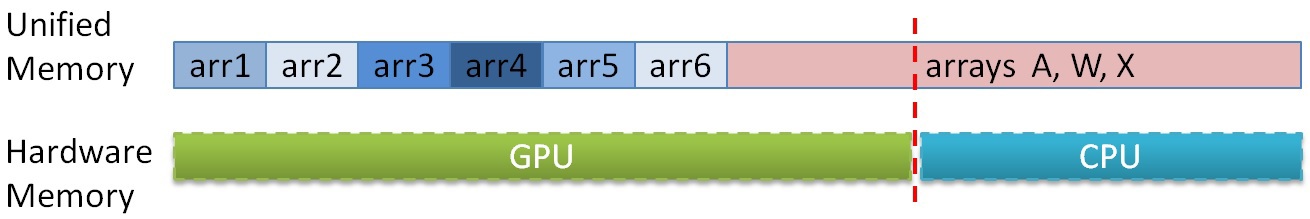
Thus, it is possible to process graphs that initially do not fit on the GPU even when using the described compression algorithms, but at a lower speed, since the throughput of PCIe is very limited.
Test results
Testing was done on the NVidia GTX Titan GPU, which has 14 SMX with 192 cuda cores (2688 in total) and a 6 cores (12th) Intel Xeon E5 v1660 processor with a frequency of 3.7 GHz. The graphs on which the testing was carried out are described above. I will give only some of the characteristics:
| Scale (2 ^ n) | Number of vertices | Number of edges (2 * M) | Count size, GB | |
|---|---|---|---|---|
| RMAT | SSCA2 | |||
| sixteen | 65,536 | 2,097,152 | ~ 2 100 000 | ~ 0.023 |
| 21 | 2,097,152 | 67 108 864 | ~ 67,200,000 | ~ 0.760 |
| 24 | 16 777 216 | 536 870 912 | ~ 537,000,000 | ~ 6.3 |
| 25 | 33,554,432 | 1,073,741,824 | ~ 1,075,000,000 | ~ 12.5 |
| 26 | 67 108 864 | 2 147 483 648 | ~ 2 150 000 000 | ~ 25.2 |
| 27 | 134 217 728 | 4,294,967,296 | ~ 4 300 000 000 | ~ 51.2 |
It can be seen that the graph of scale 16 is quite small (about 25 MB) and even without conversions it easily fits in the cache of one modern Intel Xeon processor. And since the graph weights occupy 2/3 of the total, it is actually necessary to process about 8 MB, which is only about 5 times more than the L2 cache of the GPU. However, large graphs require a sufficient amount of memory, and even graph 24 of the scale will no longer fit into the memory of the tested GPU without compression. Based on the graph representation, 26 the scale is the last one, in which the number of edges is placed in an unsigned int, which is a certain limitation of the algorithm for further scaling. This restriction is easily bypassed by expanding the data type. As it seems to me, this is not so important now, since the processing of single precision (unsigned int) is performed many times faster than double (unsigned long long) and the amount of memory is still quite small. Performance will be measured in the number of processed edges per second (traversed edges per second - TEPS).
Compilation was carried out using NVidia CUDA Toolkit 7.0 with options -O3 -arch = sm_35, Intel Composer 2015 with options -O3. The maximum performance of the implemented algorithm can be seen on the graph below:
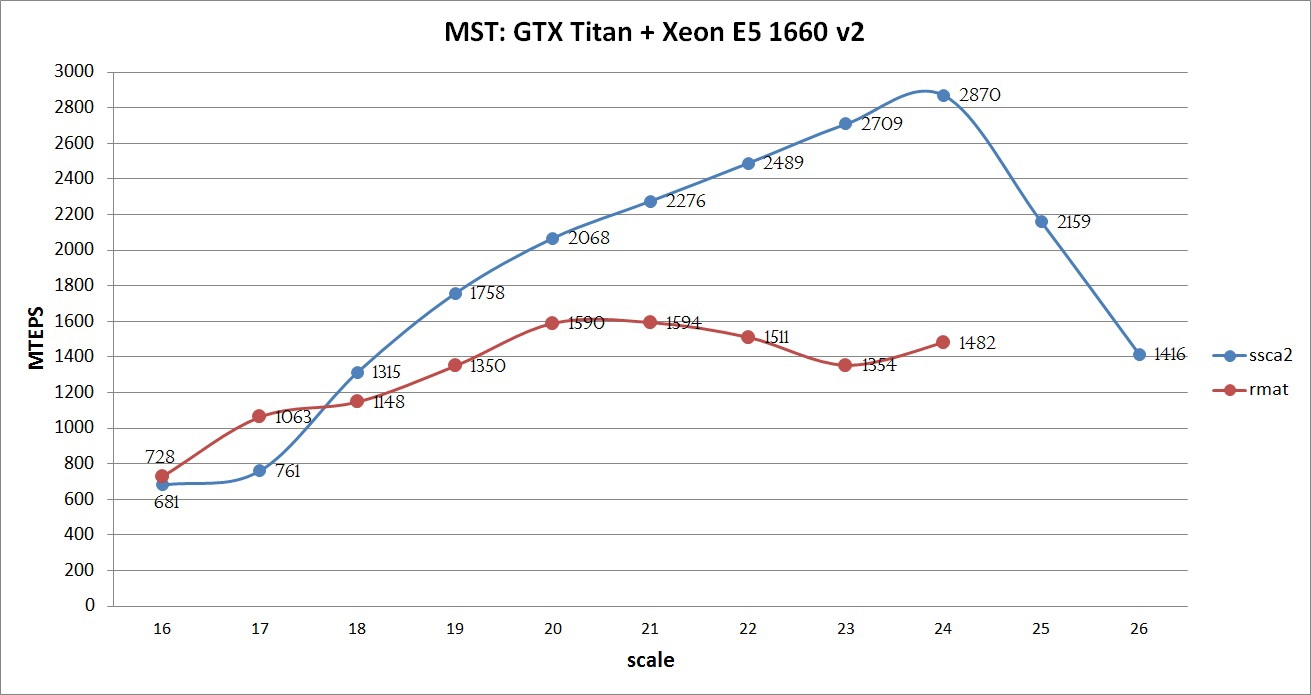
The graph shows that using all the SSCA2 optimizations, the graphs show good efficiency: the larger the graph, the better the performance. This growth is maintained until all data is stored in the memory of the GPU. On the 25th and 26th scales, the Unified Memory mechanism was used, which allowed to get the result, though with a lower speed (but as will be demonstrated below, faster than only on the CPU). If the calculation is performed on a Tesla k40 with 12GB of memory and ECC disabled and an Intel Xeon E5 V2 / V3 processor, then it would be quite possible to achieve about 3000 MTEPS on a 25 scale SSCA2 graph, and also try to process not only the 26th graph, but also 27. For the RMAT graph, such an experiment was not conducted, due to its complex structure and poor adaptation of the algorithm.
Performance comparison of different algorithms
This problem was solved within the competition of the GraphHPC 2015 conference. I would like to make a comparison with the program written by Alexander Darin, who, according to the authors, took the first place in this competition.
Since the general table contains the results on the test platform provided by the authors, it would not be superfluous to bring graphics on the CPU and GPU on the described platform (GTX Titan + Xeon E5 v2). Below are the results for two graphs:


The graphs show that the algorithm described in this article is more optimized for SSCA2 graphs, while the algorithm implemented by Alexander Daryin is well optimized for RMAT graphs. In this case, we cannot say for sure which of the implementations is the best, because each has its own advantages and disadvantages. The criterion by which algorithms must be evaluated is also unclear. If we talk about the processing of large graphs, then the fact that the algorithm can process graphs 24-26 of the scale is a big plus and advantage. If we talk about the average processing speed of graphs of any size, it is not clear what exactly the average value to consider. One thing is clear - one algorithm handles SSCA2 graphs well, the second - RMAT. If we combine these two implementations, then the average performance will be about 3200 MTEPS for scale 23. Presentation of the description of some optimizations of the algorithm of Alexander Daryin can be viewed here .
Of the foreign articles are the following.
1) [13] From this article, some ideas were used in the implementation of the described algorithm. Directly compare the results obtained by the authors can not, because testing was performed on the old NVidia Tesla S1070. The performance achieved by the authors on the GPU ranges from 18-36 MTEPS. Published in 2009 and in 2013.
2) [14] implementation of the Prima algorithm on the GPU.
3) [15] implementation of k NN-Boruvka on the GPU.
There are also some parallel implementations on the CPU. But I could not find high performance in foreign articles. Maybe someone from readers can tell if I missed something. It is also worth noting that there are practically no publications on this topic in Russia (with the exception of Vadim Zaitsev ), which is very sad, I think.
About the competition and instead of the conclusion
I would like to write my opinion on the past and mentioned competition for the best implementation of MST. These comments are not required to read and express my personal opinion. It is possible that someone thinks very differently.
The basis for solving this problem for all participants was laid, in fact, the same Boruvka algorithm. It turns out that the task is slightly simplified, since other algorithms (Kruskal and Prima) have a large computational complexity and are slow or poorly mapped to parallel architectures, including the GPU. From the conference title, it logically follows that it is necessary to write an algorithm that handles large graphs well, such graphs that, say, occupy 1GB in memory and above (such graphs had scales of 22 or more). Unfortunately, the authors for some reason did not take into account this fact and the whole competition was reduced to writing an algorithm that works well in the cache, since the test platform contained 2 CPUs with a total cache of 50 MB (graphs up to 17 scales weigh <= 50MB). Only one of the participants showed an acceptable result - Vadim Zaitsev, who received a fairly high average value on 2 CPUs on a graph of 22 scales. But as it turned out during the conference, this participant was engaged in the MST task for quite a long time. Probably, the processing speed of large graphs of the rest of the implemented algorithms will not be large and will be very different from those figures (for the worse), published on the competition website. You should also pay attention to the fact that the structures of graphs are very different and why suddenly it is necessary to consider the average performance just as the arithmetic average is also not entirely clear. The size of the processed graph was also not taken into account. Another unpleasant "feature" of the provided system (which included 2x Intel Xeon E5-2690 and NVidia Tesla K20x) is not working PCIe 3.0 (although supported on the GPU and present on the server board). As a result, it was not possible to use two faster (albeit slightly) than Xeon E5 processors, since the PCIe 2.0 speed is almost 3 times lower.
It should be noted that the solution of such problems on the GPU is not easy, since the processing of graphs is difficult to parallelize on the GPU due to its architectural features. And it is quite possible, these competitions should contribute to the development of specialists in the field of writing algorithms that use non-structural grids for graphics processors. But judging by the results of this year, as well as the previous one, the use of the GPU in such tasks, unfortunately, is very limited.
References:
[1] en.wikipedia.org/wiki/Sparse_matrix
[2] www.dislab.org/GraphHPC-2014/rmat-siam04.pdf
[3] www.dislab.org/GraphHPC-2015/SSCA2-TechReport.pdf
[4] www.nvidia.ru/object/tesla-supercomputer-workstations-ru.html
[5] www.nvidia.ru/object/geforce-gtx-titan-x-ru.html#pdpContent=2
[6] www.amd.com/ru-ru/products/graphics/workstation/firepro-3d/9100
[7] en.wikipedia.org/wiki/Bor%C5%AFvka 's_algorithm
[8] www.cs.princeton.edu/~rs/AlgsDS07/01UnionFind.pdf
[9] habrahabr.ru/company/epam_systems/blog/247805
[10] on-demand.gputechconf.com/gtc/2013/presentations/S3174-Kepler-Shuffle-Tips-Tricks.pdf
[11] openmp.org/wp
[12] devblogs.nvidia.com/parallelforall/unified-memory-in-cuda-6
[13] stanford.edu/~vibhavv/papers/old/Vibhav09Fast.pdf
[14] ieeexplore.ieee.org/xpl/login.jsp?tp=&arnumber=5678261&url=http%3A%2F%2Fieeexplore.ieee.org%2Fxpls%2Fabs_all.jsp%3Farnumber%3D5678261
[15] link.springer.com/chapter/10.1007%2F978-3-642-31125-3_6
Source: https://habr.com/ru/post/253031/
All Articles