Probabilistic Models: Sampling
Hello again! Today, I continue a series of articles in the Surfingbird blog devoted to different methods of recommendations, and also sometimes just different probabilistic models. Long ago, it seems, last Friday last summer, I wrote a small cycle about graphical probabilistic models: the first part introduced the basics of graphical probabilistic models, in the second part there were several examples, part 3 talked about the message passing algorithm, and in the fourth part we We talked briefly about variational approximations. The cycle ended with a promise to talk about sampling - well, in less than a year. Generally speaking, in this mini-cycle, I’ll talk more about the LDA model and how it helps us make textual recommendations. But today I will start with the fact that I will fulfill the long-held promise and tell you about sampling in probabilistic models - one of the main methods of approximate inference.

First of all, I remind you of what we are talking about. One of the main tools of machine learning is the Bayes theorem:
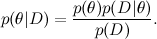
Here D is the data, θ is the model parameters that we want to train, and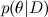 - this distribution θ, subject to the available data D ; about the Bayes theorem, we have already spoken in detail to one of the previous articles . In machine learning, we usually want to find the posterior distribution
- this distribution θ, subject to the available data D ; about the Bayes theorem, we have already spoken in detail to one of the previous articles . In machine learning, we usually want to find the posterior distribution 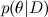 and then predict new variable values
and then predict new variable values  . In complex graphical models, all this, as we discussed in the previous series, usually comes down to the fact that we have a large and confusing distribution of various random variables.
. In complex graphical models, all this, as we discussed in the previous series, usually comes down to the fact that we have a large and confusing distribution of various random variables.  which is decomposed into a product of distributions easier, and our task is to marginalize , i.e. Sum by part of the variables or find the expectation of a function from the part of the variables. Note that all of our tasks in one way or another boil down to calculating the expectation of different functions under the condition of a complex distribution, usually a conditional distribution of a model into which the values of some variables are substituted. We can also assume that we can count the value of the joint distribution at any point of it - this usually follows easily from the general form of the model, and the difficulty lies precisely in summing-integrating along the heap of these variables.
which is decomposed into a product of distributions easier, and our task is to marginalize , i.e. Sum by part of the variables or find the expectation of a function from the part of the variables. Note that all of our tasks in one way or another boil down to calculating the expectation of different functions under the condition of a complex distribution, usually a conditional distribution of a model into which the values of some variables are substituted. We can also assume that we can count the value of the joint distribution at any point of it - this usually follows easily from the general form of the model, and the difficulty lies precisely in summing-integrating along the heap of these variables.
We already know that it is difficult to carry out an exact conclusion, and effective algorithms are obtained only for the case of a factor graph without cycles or with small cycles. However, real models often contain a bunch of variables that are tightly connected to each other and form a mass of cycles. We talked a little about variational approximations - one possible way to overcome the difficulties that arise. And today I will talk about another, even more popular method - about sampling.
')
The idea is simple to the extreme. We have already found out that, in principle, everything that we need is expressed in the form of expectations of various functions in our complex and confusing distribution of p ( x ). How to calculate the waiting function by the distribution? If we can sample from this distribution, i.e. take random points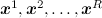 over the distribution p ( x ), then the expectation of any function can be approximated as the arithmetic average at the sampled points:
over the distribution p ( x ), then the expectation of any function can be approximated as the arithmetic average at the sampled points:

For example, to calculate the average (expected value), you need to average over the samples x .
Thus, our entire task actually comes down to learning how to sample points from a distribution, provided that we can read the value of this distribution at any point.
A person brought up on the functions of one variable (that is, in fact, anyone who has listened to the basic course of mathematical analysis at a university), it may seem that there is no big problem here, and all this talk about anything is well known, as approximately calculate the integral, you just need to break the segment into parts, count the function in the middle of each small segment, rectangles there, trapezium, you know ... And indeed, there is nothing complicated in sampling from distributions from one variable. Difficulties, as always, begin with the growth of dimensionality - if in dimension 1 you need to count 10 function values to divide the interval into segments with a side of 0.1, then in dimension 100 such values you will need 10,100 , which is much sadder. This and other effects that make work in a high dimension completely different from the usual dimension of 2-3, are called “curse of dimensionality”; there are other interesting counterintuitive effects there, it may be worth it sometime to talk about this in more detail.
There is one more difficulty - I wrote above that we can read the value of the distribution at any point. In fact, usually we can consider not the value of the distribution p ( x ), but the value of some function p * ( x ), which is proportional to p ( x ). Recall the Bayes theorem - from it we get not the a posteriori distribution itself, but the function proportional to it:
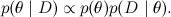
Usually at this point we said that this is sufficient, and the exact probability value can be simply calculated by normalizing the resulting distribution so that it is summed to one (that is, by calculating the denominator in the Bayes theorem). However, this is precisely the difficult task that we are now trying to learn how to solve: to sum up all the values of a complex distribution. Therefore, in real life, we cannot rely on the value of the true probabilities, only on some function proportional to them p * .
Despite all the above difficulties, sampling is still possible, and this is indeed one of the main methods of approximate inference. Moreover, as we will soon see, sampling is not as difficult as it seems. In this section, I will talk about the basic idea, on which one way or another, all the sampling algorithms are based. The idea is this: if you uniformly select a point under the graph of the density function of the distribution p , then its X-coordinate will be taken just according to the distribution p . It is very easy to understand intuitively: imagine a density graph and break it into small bars, like this (charts are made in matplotlib):
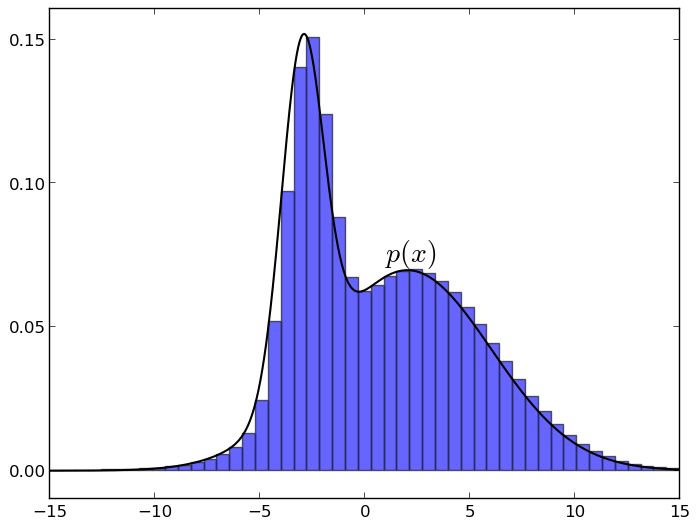
Now you can see that when you take a random point under the graph, each X-coordinate will meet with a probability proportional to the height of its column, i.e. just with the value of the density function on this column.
Thus, we only need to learn how to take a random point under the density graph of the desired distribution. How to do it?
The first idea that really works and is often applied in practice is the so-called rejection sampling . Let's assume that we have some other distribution q (more precisely, the function q * proportional to it), which we are already able to sample. Usually this is some kind of classical distribution - uniform or normal. It is worth noting here that sampling from a normal distribution is also not a completely trivial task, but we will skip it now, for us it is enough to know that “people can do this”. Further, suppose that we know a constant c about it such that for all x . Then we can sample p in this way:
. Then we can sample p in this way:
Here is a picture illustrating a sample with a deviation (normal distribution, chosen to majorize a mixture of two other Gaussians):
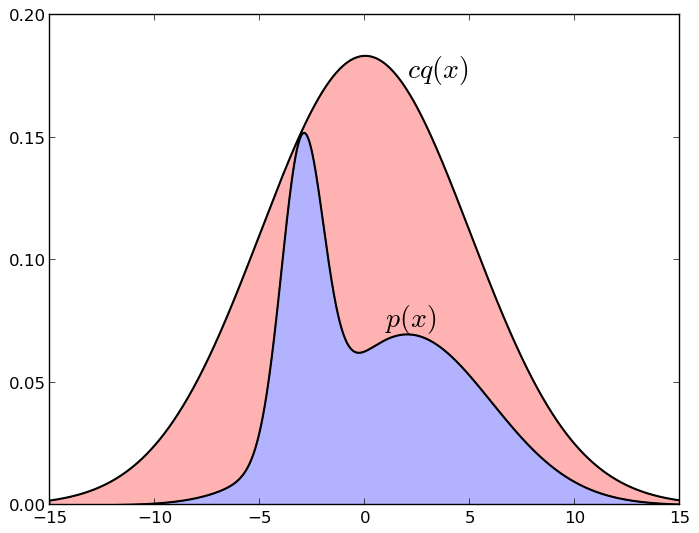
If a point under the graph cq * falls under the graph p * , i.e. in the blue zone, we take it; and if above it, i.e. in the red zone, - do not take. Again, it is easy to understand intuitively why it works: we take a random point evenly under the cq * schedule, and then leave only those points that fall under the p * schedule - it is clear that we will have just the uniform distribution under schedule p * . It is also clear that if we manage to choose q sufficiently similar to p (for example, we know approximately where p is concentrated and cover this area with a Gaussian), then this method will be much more efficient than simply breaking the whole space into equal pieces. But in order for the method to work, it is necessary that cq really approximates p quite well - if the area of the blue zone is 10 -10 from the area of red, no samples will work ...
There are different versions of this idea, which I will not dwell on in detail. In short, this idea and its variations work well with complex densities in dimensions, measured in units, but in a really high dimension, complexity begins. Because of the effects of cursing the dimension, classical test distributions q get different unpleasant properties (for example, in high dimension the orange peel occupies almost its entire volume, and, in particular, the normal distribution is also almost entirely concentrated in a very thin “peel”, and not at all zero), samples in the right places can not be obtained, and the whole thing often fails. Other methods are needed.
The essence of this algorithm is based on the same idea: we want to take the point uniformly under the graph of the function. However, the approach is now different: instead of trying to cover the entire distribution with the “cap” of the function q , we will act from the inside: we will build a random walk under the graph of the function, moving from one point to another, and from time to time take the current point of the walk as a sample. Since in this case this random walk is a Markov chain (i.e. its next point depends only on the previous one, but there is no memory), this class of algorithms is also called MCMC sampling (Markov chain Monte Carlo). To prove that the Metropolis-Hastings algorithm really works, you need to know the properties of Markov chains; but we are not going to prove anything formally now, so I’ll just chastely leave the link to the wiki .
In fact, the algorithm is similar to the same sample with a deviation, but with an important difference: before, the distribution of q was the same for the whole space, and now it will be local and will change with time, depending on the current wandering point. As before, we consider the family of test distributions.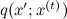 where
where 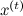 - Current state. But now q should not be an approximation of p , but should simply be some sampled distribution that is concentrated in the vicinity of the current point — for example, a spherical Gaussian with a small dispersion is well suited. The candidate for the new state x 'will now be sampled from
- Current state. But now q should not be an approximation of p , but should simply be some sampled distribution that is concentrated in the vicinity of the current point — for example, a spherical Gaussian with a small dispersion is well suited. The candidate for the new state x 'will now be sampled from 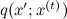 . The next iteration of the algorithm begins with the state
. The next iteration of the algorithm begins with the state 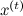 and is this:
and is this:
The point is that we move to a new distribution center, if we take the next step, and we get a random walk depending on the distribution p : we always take a step, if the density function increases in this direction, and sometimes we reject it if it decreases (we do not always reject , because we need to be able to go out of local maxima and wander under the whole schedule p ). I note, by the way, that when a step is rejected, we do not just discard the new point, but repeat the same time x ( t ) a second time - thus there is a high probability of repeating samples around the local maxima of the distribution p , which corresponds to a higher density.
And again the picture is a typical walk on a plane under the graph of a mixture of two two-dimensional Gaussians:
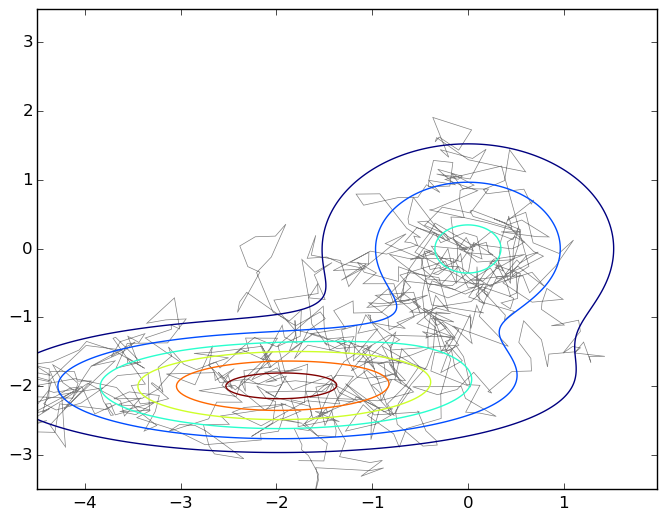
To emphasize how simple this algorithm really is, I will give the code that generated this graph.
So, we got a random walk under the density graph p ( x ); in fact, it is still necessary to prove it, but we will not go deep into mathematics, so take my word for it. What now to do with this random walk, we also need independent samples, and here it turns out that the next point of the walk obviously lies next to the previous one, and there is no independence at all?
The answer is simple: you need to take not every point, but only some, separated from each other by many steps. Since this is a random walk, then if most of q is concentrated in radius ε, and the total radius in which p is concentrated, is equal to D , then to obtain an independent sample, you will need about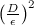 steps. In this you, again, can take a word, or you can recall the classic puzzle from the course of probability theory about how far in n steps the point goes, which moves with equal probability to the left and to the right; it will go on average
steps. In this you, again, can take a word, or you can recall the classic puzzle from the course of probability theory about how far in n steps the point goes, which moves with equal probability to the left and to the right; it will go on average  , so it is logical that the steps will be a quadratic number.
, so it is logical that the steps will be a quadratic number.
And now the main feature: this quadratic number depends on the radii of the two distributions, but it is completely independent of the dimension . Monte Carlo Markov chain sampling is much less subject to curse dimension than the other methods we discussed; that is why it has become, one might say, the gold standard. In practice, however, it is difficult to estimate D , so in concrete implementations they usually do this: first, quite a few number of first steps, say 1000, are discarded (this is called burn-in, and this is really important because the starting point can fall into an unsuccessful area p , so that before starting the process, you need to make sure that we have already walked for a long time), and then take each nth sample, where n is selected experimentally, based on the actual autocorrelation in the sample sequence.
And the latest sampling algorithm for today is the Gibbs sampling. This is actually a special case of the Metropolis-Hastings algorithm, a very popular and simple special case.
The idea of sampling according to Gibbs is quite simple: suppose that we are in a very large dimension, the vector x is very long, and it is difficult for us to choose the entire sample at once, it does not work. Let's try to choose a sample not all at once, but component by component. Then surely these one-dimensional distributions will be simpler, and we will choose the sample.
Formally speaking, we run through the components of the vector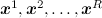 , at each step, we fix all the variables, except for one, and choose sequentially
, at each step, we fix all the variables, except for one, and choose sequentially 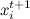 by distribution
by distribution
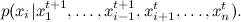
This differs from the general case of wandering around Metropolis-Hastings in that now we move at each step along one of the coordinate axes; Here is the corresponding picture:
Gibbs sampling is a special case of the Metropolis algorithm for distributions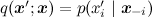 , and the probability of taking each sample is always equal to 1 (you can prove it as an exercise). Therefore, Gibbs sampling converges, and since this is the same random walk in essence, the same quadratic estimate is true.
, and the probability of taking each sample is always equal to 1 (you can prove it as an exercise). Therefore, Gibbs sampling converges, and since this is the same random walk in essence, the same quadratic estimate is true.
Gibbs sampling does not require any special assumptions or knowledge. You can quickly make a working model, and therefore it is a very popular algorithm. We also note that in large dimensions it may be more efficient to sample several variables at once, rather than one — for example, it often happens that we have a bipartite graph from variables in which all the variables from one fraction are associated with all the variables from another fraction (well or with many), and are not related to each other. In such a situation, God himself ordered to fix all the variables of one beat and sample all the variables in the other beat at the same time (this can be taken literally - since with this structure all variables of one beat are conditionally independent under the condition of the other, they can be sampled independently and in parallel), then fix all variables of the second beat and so on.
Today we managed quite a lot: we talked about different sampling methods, which is one of the main tools for approximate inference in complex probabilistic models. Next, I plan to develop this new mini-cycle in the direction of thematic modeling, more precisely, the model LDA (latent Dirichlet allocation, latent placement of Dirichlet). In the next series I will talk about the model itself (I have already described it briefly, now you can talk in more detail) and explain how the Gibbs sampling works for LDA. And then we will gradually move on to how LDA and its numerous extensions can be applied to recommender systems.

What is the problem
First of all, I remind you of what we are talking about. One of the main tools of machine learning is the Bayes theorem:
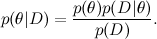
Here D is the data, θ is the model parameters that we want to train, and
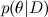 - this distribution θ, subject to the available data D ; about the Bayes theorem, we have already spoken in detail to one of the previous articles . In machine learning, we usually want to find the posterior distribution
- this distribution θ, subject to the available data D ; about the Bayes theorem, we have already spoken in detail to one of the previous articles . In machine learning, we usually want to find the posterior distribution 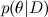 and then predict new variable values
and then predict new variable values  . In complex graphical models, all this, as we discussed in the previous series, usually comes down to the fact that we have a large and confusing distribution of various random variables.
. In complex graphical models, all this, as we discussed in the previous series, usually comes down to the fact that we have a large and confusing distribution of various random variables.  which is decomposed into a product of distributions easier, and our task is to marginalize , i.e. Sum by part of the variables or find the expectation of a function from the part of the variables. Note that all of our tasks in one way or another boil down to calculating the expectation of different functions under the condition of a complex distribution, usually a conditional distribution of a model into which the values of some variables are substituted. We can also assume that we can count the value of the joint distribution at any point of it - this usually follows easily from the general form of the model, and the difficulty lies precisely in summing-integrating along the heap of these variables.
which is decomposed into a product of distributions easier, and our task is to marginalize , i.e. Sum by part of the variables or find the expectation of a function from the part of the variables. Note that all of our tasks in one way or another boil down to calculating the expectation of different functions under the condition of a complex distribution, usually a conditional distribution of a model into which the values of some variables are substituted. We can also assume that we can count the value of the joint distribution at any point of it - this usually follows easily from the general form of the model, and the difficulty lies precisely in summing-integrating along the heap of these variables.We already know that it is difficult to carry out an exact conclusion, and effective algorithms are obtained only for the case of a factor graph without cycles or with small cycles. However, real models often contain a bunch of variables that are tightly connected to each other and form a mass of cycles. We talked a little about variational approximations - one possible way to overcome the difficulties that arise. And today I will talk about another, even more popular method - about sampling.
')
Monte Carlo approach
The idea is simple to the extreme. We have already found out that, in principle, everything that we need is expressed in the form of expectations of various functions in our complex and confusing distribution of p ( x ). How to calculate the waiting function by the distribution? If we can sample from this distribution, i.e. take random points
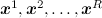 over the distribution p ( x ), then the expectation of any function can be approximated as the arithmetic average at the sampled points:
over the distribution p ( x ), then the expectation of any function can be approximated as the arithmetic average at the sampled points:
For example, to calculate the average (expected value), you need to average over the samples x .
Thus, our entire task actually comes down to learning how to sample points from a distribution, provided that we can read the value of this distribution at any point.
What is the difficulty
A person brought up on the functions of one variable (that is, in fact, anyone who has listened to the basic course of mathematical analysis at a university), it may seem that there is no big problem here, and all this talk about anything is well known, as approximately calculate the integral, you just need to break the segment into parts, count the function in the middle of each small segment, rectangles there, trapezium, you know ... And indeed, there is nothing complicated in sampling from distributions from one variable. Difficulties, as always, begin with the growth of dimensionality - if in dimension 1 you need to count 10 function values to divide the interval into segments with a side of 0.1, then in dimension 100 such values you will need 10,100 , which is much sadder. This and other effects that make work in a high dimension completely different from the usual dimension of 2-3, are called “curse of dimensionality”; there are other interesting counterintuitive effects there, it may be worth it sometime to talk about this in more detail.
There is one more difficulty - I wrote above that we can read the value of the distribution at any point. In fact, usually we can consider not the value of the distribution p ( x ), but the value of some function p * ( x ), which is proportional to p ( x ). Recall the Bayes theorem - from it we get not the a posteriori distribution itself, but the function proportional to it:
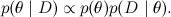
Usually at this point we said that this is sufficient, and the exact probability value can be simply calculated by normalizing the resulting distribution so that it is summed to one (that is, by calculating the denominator in the Bayes theorem). However, this is precisely the difficult task that we are now trying to learn how to solve: to sum up all the values of a complex distribution. Therefore, in real life, we cannot rely on the value of the true probabilities, only on some function proportional to them p * .
What to do: we sample under the schedule
Despite all the above difficulties, sampling is still possible, and this is indeed one of the main methods of approximate inference. Moreover, as we will soon see, sampling is not as difficult as it seems. In this section, I will talk about the basic idea, on which one way or another, all the sampling algorithms are based. The idea is this: if you uniformly select a point under the graph of the density function of the distribution p , then its X-coordinate will be taken just according to the distribution p . It is very easy to understand intuitively: imagine a density graph and break it into small bars, like this (charts are made in matplotlib):

Now you can see that when you take a random point under the graph, each X-coordinate will meet with a probability proportional to the height of its column, i.e. just with the value of the density function on this column.
Thus, we only need to learn how to take a random point under the density graph of the desired distribution. How to do it?
The first idea that really works and is often applied in practice is the so-called rejection sampling . Let's assume that we have some other distribution q (more precisely, the function q * proportional to it), which we are already able to sample. Usually this is some kind of classical distribution - uniform or normal. It is worth noting here that sampling from a normal distribution is also not a completely trivial task, but we will skip it now, for us it is enough to know that “people can do this”. Further, suppose that we know a constant c about it such that for all x
 . Then we can sample p in this way:
. Then we can sample p in this way:- we take sample x on distribution q * ( x );
- take a random number u uniformly from the interval
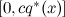 ;
; - calculate p * ( x ); if it is greater than u , then x is added to the samples, and if it is less (that is, if u does not fall under the density curve p * ), then x is rejected (hence the sample with the deviation).
Here is a picture illustrating a sample with a deviation (normal distribution, chosen to majorize a mixture of two other Gaussians):

If a point under the graph cq * falls under the graph p * , i.e. in the blue zone, we take it; and if above it, i.e. in the red zone, - do not take. Again, it is easy to understand intuitively why it works: we take a random point evenly under the cq * schedule, and then leave only those points that fall under the p * schedule - it is clear that we will have just the uniform distribution under schedule p * . It is also clear that if we manage to choose q sufficiently similar to p (for example, we know approximately where p is concentrated and cover this area with a Gaussian), then this method will be much more efficient than simply breaking the whole space into equal pieces. But in order for the method to work, it is necessary that cq really approximates p quite well - if the area of the blue zone is 10 -10 from the area of red, no samples will work ...
There are different versions of this idea, which I will not dwell on in detail. In short, this idea and its variations work well with complex densities in dimensions, measured in units, but in a really high dimension, complexity begins. Because of the effects of cursing the dimension, classical test distributions q get different unpleasant properties (for example, in high dimension the orange peel occupies almost its entire volume, and, in particular, the normal distribution is also almost entirely concentrated in a very thin “peel”, and not at all zero), samples in the right places can not be obtained, and the whole thing often fails. Other methods are needed.
Metropolis-Hastings Algorithm
The essence of this algorithm is based on the same idea: we want to take the point uniformly under the graph of the function. However, the approach is now different: instead of trying to cover the entire distribution with the “cap” of the function q , we will act from the inside: we will build a random walk under the graph of the function, moving from one point to another, and from time to time take the current point of the walk as a sample. Since in this case this random walk is a Markov chain (i.e. its next point depends only on the previous one, but there is no memory), this class of algorithms is also called MCMC sampling (Markov chain Monte Carlo). To prove that the Metropolis-Hastings algorithm really works, you need to know the properties of Markov chains; but we are not going to prove anything formally now, so I’ll just chastely leave the link to the wiki .
In fact, the algorithm is similar to the same sample with a deviation, but with an important difference: before, the distribution of q was the same for the whole space, and now it will be local and will change with time, depending on the current wandering point. As before, we consider the family of test distributions.
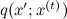 where
where 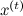 - Current state. But now q should not be an approximation of p , but should simply be some sampled distribution that is concentrated in the vicinity of the current point — for example, a spherical Gaussian with a small dispersion is well suited. The candidate for the new state x 'will now be sampled from
- Current state. But now q should not be an approximation of p , but should simply be some sampled distribution that is concentrated in the vicinity of the current point — for example, a spherical Gaussian with a small dispersion is well suited. The candidate for the new state x 'will now be sampled from 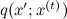 . The next iteration of the algorithm begins with the state
. The next iteration of the algorithm begins with the state 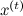 and is this:
and is this:- select x 'by distribution
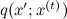 ;
; - calculate

(don't be scared, for symmetric distributions, for example, Gaussian, equals 1, this is just an asymmetry correction);
for symmetric distributions, for example, Gaussian, equals 1, this is just an asymmetry correction); - With probability a (1, if a > 1)
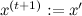 otherwise
otherwise 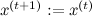 .
.
The point is that we move to a new distribution center, if we take the next step, and we get a random walk depending on the distribution p : we always take a step, if the density function increases in this direction, and sometimes we reject it if it decreases (we do not always reject , because we need to be able to go out of local maxima and wander under the whole schedule p ). I note, by the way, that when a step is rejected, we do not just discard the new point, but repeat the same time x ( t ) a second time - thus there is a high probability of repeating samples around the local maxima of the distribution p , which corresponds to a higher density.
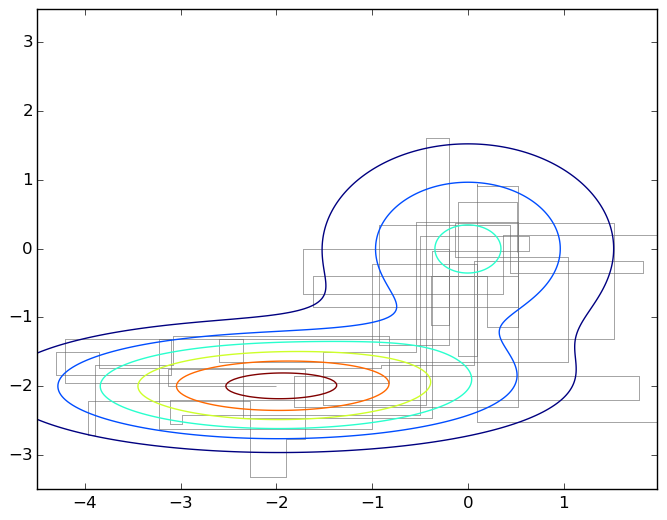
And again the picture is a typical walk on a plane under the graph of a mixture of two two-dimensional Gaussians:

To emphasize how simple this algorithm really is, I will give the code that generated this graph.
Python code
import numpy as np import matplotlib.pyplot as plt import matplotlib.patches as patches import matplotlib.path as path import matplotlib.mlab as mlab import scipy.stats as stats delta = 0.025 X, Y = np.meshgrid(np.arange(-4.5, 2.0, delta), np.arange(-3.5, 3.5, delta)) z1 = stats.multivariate_normal([0,0],[[1.0,0],[0,1.0]]) z2 = stats.multivariate_normal([-2,-2],[[1.5,0],[0,0.5]]) def z(x): return 0.4*z1.pdf(x) + 0.6*z2.pdf(x) Z1 = mlab.bivariate_normal(X, Y, 1.0, 1.0, 0.0, 0.0) Z2 = mlab.bivariate_normal(X, Y, 1.5, 0.5, -2, -2) Z = 0.4*Z1 + 0.6*Z2 Q = stats.multivariate_normal([0,0],[[0.05,0],[0,0.05]]) r = [0,0] samples = [r] for i in range(0,1000): rq = Q.rvs() + r a = z(rq)/z(r) if np.random.binomial(1,min(a,1),1)[0] == 1: r = rq samples.append(r) codes = np.ones(len(samples), int) * path.Path.LINETO codes[0] = path.Path.MOVETO p = path.Path(samples,codes) fig, ax = plt.subplots() ax.contour(X, Y, Z) ax.add_patch(patches.PathPatch(p, facecolor='none', lw=0.5, edgecolor='gray')) plt.show() So, we got a random walk under the density graph p ( x ); in fact, it is still necessary to prove it, but we will not go deep into mathematics, so take my word for it. What now to do with this random walk, we also need independent samples, and here it turns out that the next point of the walk obviously lies next to the previous one, and there is no independence at all?
The answer is simple: you need to take not every point, but only some, separated from each other by many steps. Since this is a random walk, then if most of q is concentrated in radius ε, and the total radius in which p is concentrated, is equal to D , then to obtain an independent sample, you will need about
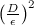 steps. In this you, again, can take a word, or you can recall the classic puzzle from the course of probability theory about how far in n steps the point goes, which moves with equal probability to the left and to the right; it will go on average
steps. In this you, again, can take a word, or you can recall the classic puzzle from the course of probability theory about how far in n steps the point goes, which moves with equal probability to the left and to the right; it will go on average  , so it is logical that the steps will be a quadratic number.
, so it is logical that the steps will be a quadratic number.And now the main feature: this quadratic number depends on the radii of the two distributions, but it is completely independent of the dimension . Monte Carlo Markov chain sampling is much less subject to curse dimension than the other methods we discussed; that is why it has become, one might say, the gold standard. In practice, however, it is difficult to estimate D , so in concrete implementations they usually do this: first, quite a few number of first steps, say 1000, are discarded (this is called burn-in, and this is really important because the starting point can fall into an unsuccessful area p , so that before starting the process, you need to make sure that we have already walked for a long time), and then take each nth sample, where n is selected experimentally, based on the actual autocorrelation in the sample sequence.
Gibbs sampling
And the latest sampling algorithm for today is the Gibbs sampling. This is actually a special case of the Metropolis-Hastings algorithm, a very popular and simple special case.
The idea of sampling according to Gibbs is quite simple: suppose that we are in a very large dimension, the vector x is very long, and it is difficult for us to choose the entire sample at once, it does not work. Let's try to choose a sample not all at once, but component by component. Then surely these one-dimensional distributions will be simpler, and we will choose the sample.
Formally speaking, we run through the components of the vector
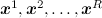 , at each step, we fix all the variables, except for one, and choose sequentially
, at each step, we fix all the variables, except for one, and choose sequentially 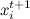 by distribution
by distribution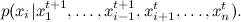
This differs from the general case of wandering around Metropolis-Hastings in that now we move at each step along one of the coordinate axes; Here is the corresponding picture:

Gibbs sampling is a special case of the Metropolis algorithm for distributions
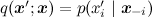 , and the probability of taking each sample is always equal to 1 (you can prove it as an exercise). Therefore, Gibbs sampling converges, and since this is the same random walk in essence, the same quadratic estimate is true.
, and the probability of taking each sample is always equal to 1 (you can prove it as an exercise). Therefore, Gibbs sampling converges, and since this is the same random walk in essence, the same quadratic estimate is true.Gibbs sampling does not require any special assumptions or knowledge. You can quickly make a working model, and therefore it is a very popular algorithm. We also note that in large dimensions it may be more efficient to sample several variables at once, rather than one — for example, it often happens that we have a bipartite graph from variables in which all the variables from one fraction are associated with all the variables from another fraction (well or with many), and are not related to each other. In such a situation, God himself ordered to fix all the variables of one beat and sample all the variables in the other beat at the same time (this can be taken literally - since with this structure all variables of one beat are conditionally independent under the condition of the other, they can be sampled independently and in parallel), then fix all variables of the second beat and so on.
Conclusion
Today we managed quite a lot: we talked about different sampling methods, which is one of the main tools for approximate inference in complex probabilistic models. Next, I plan to develop this new mini-cycle in the direction of thematic modeling, more precisely, the model LDA (latent Dirichlet allocation, latent placement of Dirichlet). In the next series I will talk about the model itself (I have already described it briefly, now you can talk in more detail) and explain how the Gibbs sampling works for LDA. And then we will gradually move on to how LDA and its numerous extensions can be applied to recommender systems.
Source: https://habr.com/ru/post/226677/
All Articles