The Human Brain Project: how do we know how the brain works?
At Habrahabr at the very beginning of 2013, after the announcement of the launch of a European mega-project to study the human brain with a budget of more than a billion euros for 10 years, a corresponding note was published. At the end of last year, the project was officially launched, and the first funds were allocated, but so far not a single word has been written about what scientific basis underlies the upcoming titanic work, comparable in importance and scale to the decoding of the human genome and manned mission to mars.
At the end of the post, you can also ask questions to the person directly working in the The Blue Brain Project team, the answers to which will be released in a separate post.
')
Instead of the preface
So, the whole mega-project is divided into 12 subprojects, which, it seems, are implemented separately - how to cross high-load computing with biology, for example - but are closely connected and intertwined. I will not tire with long and tedious stories about each of the projects. For this, there are short videos on the official The Human Brain Project Youtube channel (HBP, additional information can be found on the project’s official website ). General goals and objectives of the project are indicated in this video:
In my opinion, one of the basic, fundamental and therefore the most important subproject is the direct study of the structure of neurons, their contacts and location - the neural network - in the brain (Subproject 1 - The Mouse Brain Subproject), as well as the visualization of such networks for the subsequent construction of models and , in fact, the implementation of this knowledge is already in the gland, in calculations.
Let's start with a minute of history. The exact date of discovery of nerve cells, neurons , as such, perhaps, can not be given. One can only say with confidence that by the end of the 19th century a sufficient store of knowledge about the functioning of nervous tissue had been accumulated so that in 1906 Messrs. Golgi and Ramon and Kahal shared the Nobel Prize in Medicine for his work on the structure of the nervous system and the classification of nerve cells.
Over the past hundred years, we have understood the basic principles of the functioning of nervous tissue, learned even at a relatively primitive level to interfere with the processes occurring in it (for example, anesthesia or operations affecting the nervous tissue directly), but until now it has been a big spot on how the brain stores information as it is specifically processed by neurons, is transformed.
How to find out where and where signals go to nerve tissue?
Omitting the details of the fact that nerve cells are different, that they are connected in different ways in the nervous tissue, perform different functions, you can, however, identify common features in the structure of neurons:
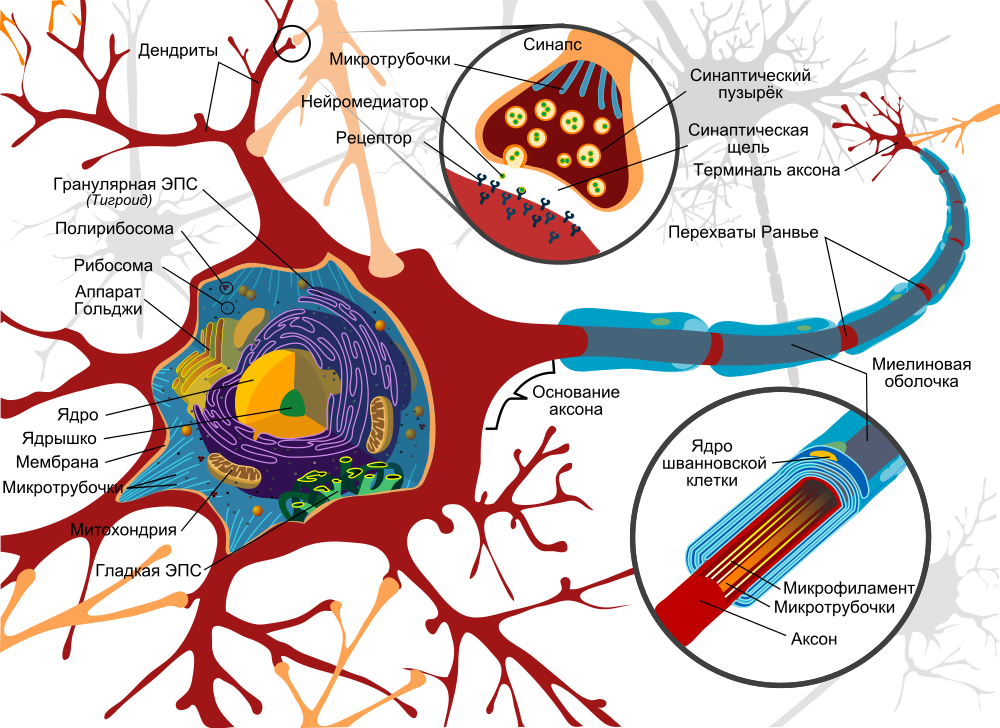
Neuron model. A source
The basis of signal transmission are axons, which, like electrical cables, stretch from one neuron to another and are necessary for the transmission of impulses. Theoretically, they can reach an enormous length - up to a meter. The same axons are attached to another neuron using a synapse, while at the end of each axon there are synaptic vesicles with neurotransmitters:
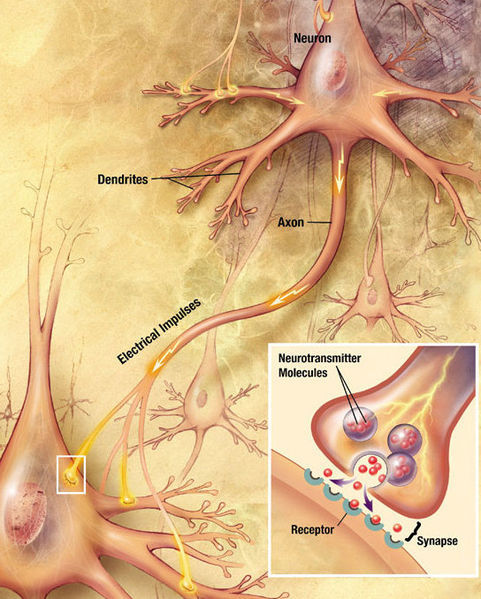
Transmission of impulses between two neurons. A source
Thus, first of all, we are interested in two things: the axon itself and the attachment point of the axon to the next nerve cell, which can be determined by synaptic vesicles with neurotransmitters. To do this, only two methods are available to us, by and large: fluorescence optical microscopy and electron microscopy.
Anticipating a logical question, why I do not consider MRI ( magnetic resonance imaging ) and functional MRI, these methods are used more to localize the areas responsible for certain functions (hearing, sight, etc.), but not to see individual neurons and their networks.
Next, I will talk about the first subproject - the study of the brain of a mouse, but not of a person (unfortunately, ethical reasons prevail over the mind). The second subproject is dedicated to the use of MRI. Thus, the study of the brain of poor mice gives us information about the fine structure of the brain, which scientists then try to associate with MRI data.
But back. In the first case - the case of fluorescence microscopy - it is necessary to paint the neurons using a fluorescent dye, and then take a sample of the sample when the dye is excited with light of a certain wavelength (often UV), but the resolution of this method will allow to see only the axons themselves, for example how they will be painted in different colors, but in general, we can hardly determine how neurons interact.
A few examples:
A new milestone - fluorescent 3D microscopy. More in Russian
A video in English about how fluorescence microscopy works in the case of nerve cells can be viewed here . Unfortunately, there is no embedded player, so click on the link, please ...
Another method is electron microscopy, the resolution of which is a few nanometers in the case of scanning and a fraction of a nanometer in the case of transmission. However, these methods are suitable either for surface analysis (scanning) or thin samples - up to 100 nm (translucent). It seems that we are at a dead end, is not it?
The hitch is aggravated by the fact that the whole tissue (brain or some other) for the most part consists of three main elements: carbon, nitrogen and oxygen, that is, light elements. Even the most advanced electron microscope in the world will not give a contrast between light elements - and yes even mixed and evenly distributed. It is not physically possible.
How long, if short ... but scientists have found a way out of this trap. What information do we want to extract? In fact, we need to know the location of the cell membranes inside the tissue, and then we could “restore” the location of the cells themselves and their “viscera. And the solution was found in the use of heavy metal salts for “tinting” the membranes. For example, it turned out that the osmium compound, OsO 4, is very well deposited on the cell membranes or, as it is commonly called, concentrated.
Actually, the matter is small - to visualize. For a long time, only transmission electron microscopy was used, requiring very thin samples, which prompted engineers to develop an ultra-cryo-microtome capable of cutting off a layer of 30-50 nm from tissue. Transmission microscopy allowed many generations of scientists to study brain slices, gave us knowledge about the internal structure of cells, was used to study the bio-tolerance of implants, but today this method has been replaced by another, scanning electron microscopy with 3D reconstruction.
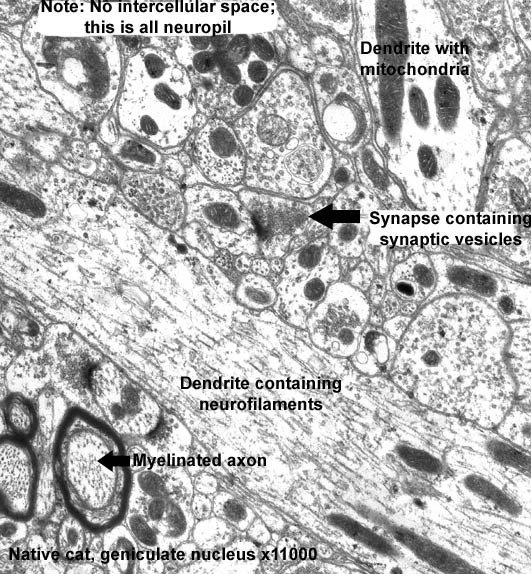
The TEM micrograph of a neuropil is an accumulation of nerve cell processes (an increase of 11,000 times). A source
3D microscopy: new features
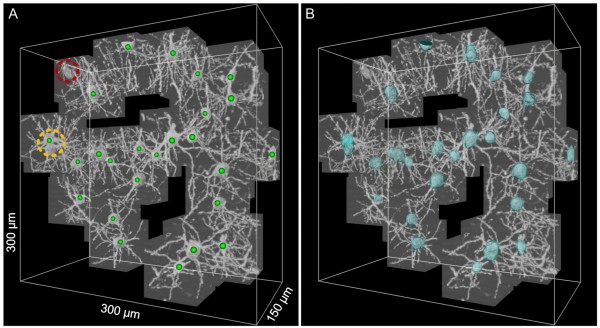
Good. In one way or another, we were able to identify a number of auxiliary cells in the chaos of nerve cells (which is really chaos, because there are a lot of auxiliary cells in the tissue itself), but from the point of view of building neural networks, this is virtually useless information, since there is no opportunities to see the location of cells in three-dimensional space, that is, information about the three-dimensional organization is hidden from us. And here, I would say, a unique method was found - three-dimensional microscopy, which became possible only in the last few years, thanks to the development of computing power of computers and processing electronics of microscopes.
At the EPFL electron microscopy center , an approach was successfully applied that combines the complete cycle of sample preparation of brain tissue and its analysis followed by 3D reconstruction. This video presents the main stages of the experiment:

The results of fixation of the sample and the replacement of water with a special resin (1:30 video): a. 80 micron mouse brain slice, bc. Cut and fixation of the area of interest 3x3 mm, df. Final treatment with a microtome.

The principle of operation of scanning electron microscopy with focused ion beam FIB / SEM (4:00 on video): ab. Schematic arrangement of the FIB-beam, cutting off a part of the tissue and the electron beam, giving the image, cd. SEM micrographs of the treated tissue.
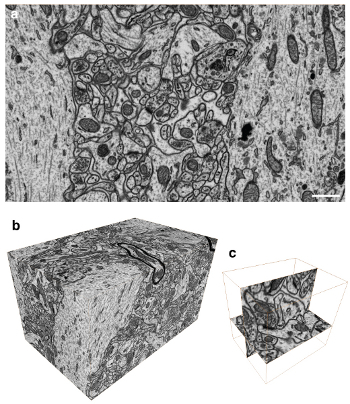
a. Visualization using FIB / SEM (5:55 video) and bc. The end result (8:00 on video)
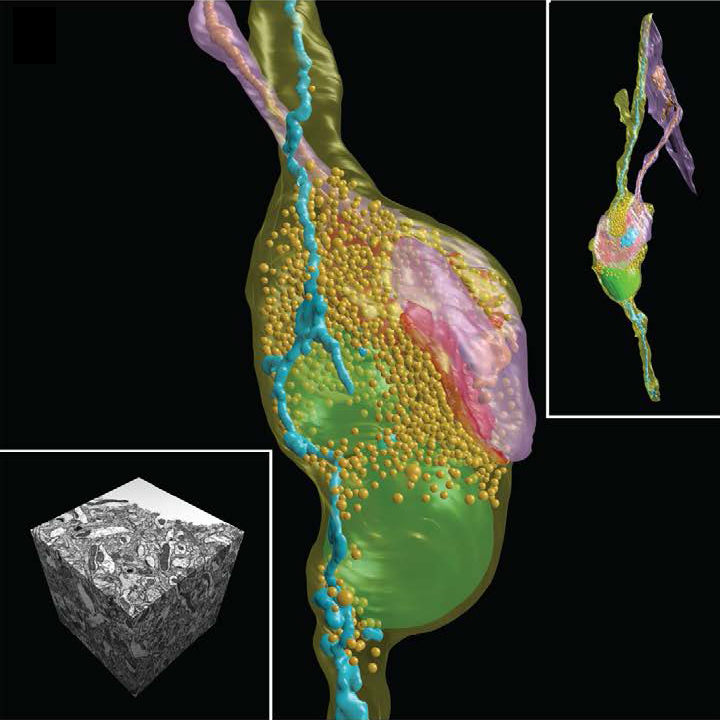
The main advantage of the method is the “cutting off” of only 5 nm layer of the brain tissue with a focused ion beam (FIB) and subsequent scanning to obtain an electron beam image (SEM). Thus, we have the opportunity to accurately study the structure of the nervous tissue, but more importantly - now the process of processing such stacks (up to 2000 images) can be performed virtually automatically without human intervention, thanks to the use of special image analysis algorithms, which often give accurate reconstruction no lower than microscopic professionals.
The group Marco Cantoni ( Marco Cantoni ) used a free-ware program specially developed for this kind of analysis - ilastik . The project has its own representation on github , so if someone from the respected Habr's readers will be interested in participating in the project or proposing their new ideas, I don’t think that the developers will refuse.
Ultimately, we have a 3D map of a piece of brain in the shape of a cube with an edge of only a few tens of microns (yes, a little, but Moscow was also not immediately built), but do not despair and give up. We got practically what we wanted - quite accurately visualized in 3D not the entire neural network, of course, but individual neurons, and one step closer to the main goal. This, according to the authors of the grandiose project, will allow a better understanding of the principles of organization and operation of the brain.
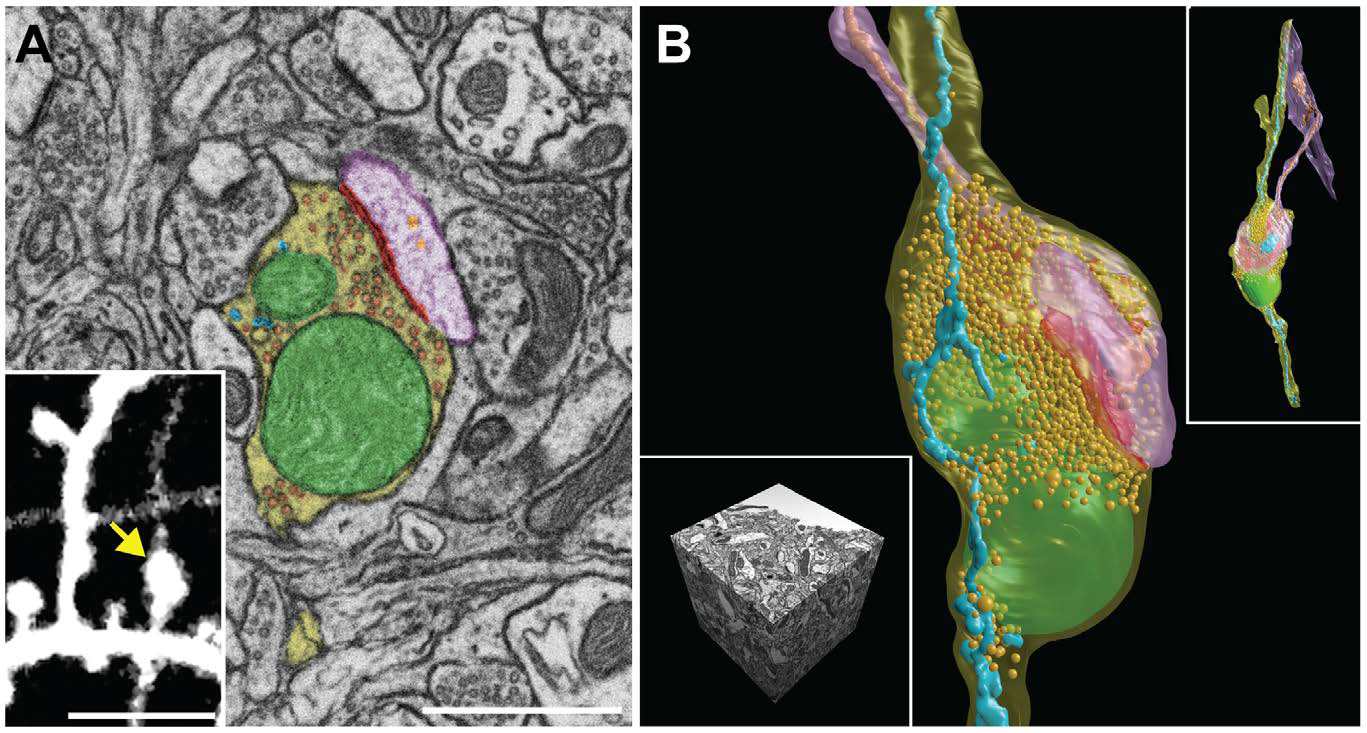
SEM micrographs and 3D reconstructions of synoptic contact and all membranes inside the mouse brain neuron. Scale A - 1 micron, inset in image A - 5 microns.
PS: Looking back and understanding what a breakthrough has been made by electron microscopy over the past decade, I would like to note that automated systems, including those developed under this project, will allow processing large data files in 5-7 years, and then it will remain for the small - only to create a 3D map of our brain or “the device, thanks to which we think what we think” (Ambrose Beers).
In preparing the materials were used open sources: 1 , 2 , 3
Sometimes briefly, and sometimes not so much about the news of science and technology, you can read on my Telegram channel - welcome;)
Source: https://habr.com/ru/post/211997/
All Articles