Shortest Common Superstring Problem
The problem of the shortest common superline is formulated as follows: to find the shortest line such that each line from the given set would be its substring. This problem occurs both in bioinformatics (the task of assembling the genome in general) and in compressing data (instead of data, store their superline and a sequence of pairs, species (index of occurrence, length)).
When I searched the network for information on this issue and its solution in Russian, there were only a couple of posts about bioinformatics, where these words are mentioned in passing. The code (except for the greedy algorithm), of course, was not there either. Having understood the problem, this fact was inspired by the article here.
Be careful, 4 megabytes!
For the most part, the article is an attempt to translate into understandable language, illustrate, spice up with an example, and, of course, implement a 4-approximate algorithm for constructing superstructures from Dan Gasfield's book (see the used literature). But first, a small introduction and a 2-approximate, greedy algorithm.
')
In the simplest case (if the name of the problem did not include the word “shortest”), the solution of the problem would be a simple concatenation of the input strings in an arbitrary order. But in this formulation of the problem, we are dealing with NP-completeness, which means that at present there is no algorithm by which one could solve this problem on a Turing machine in a time that does not exceed the polynomial of the data size.
Input: S 1 , S 2 , ..., S n - the set of lines of a finite alphabet E *.
Output: X is the string of the alphabet E * containing each string S 1..n as a substring, where the length | X | minimized.
Example. Take the set of lines S = {abc, cde, eab}. In the case of concatenation, the length of the output superline will be 9 (X = abccdeeab), which is obviously not the best option, since the lines can have a suffix-prefix match. We will call the length of this coincidence overlap . (The choice of an English word is not accidental, since it does not have a specific and unambiguous translation into Russian. In the Russian-language literature, the terms overlay, intersection, overlap, and suffix-prefix coincidence are commonly used).
Retreat. The concept of overlap is one of the most important in the implementation process of the construction of superstructures. Calculating overlap for an ordered pair of strings (S i , S j ) is reduced to finding the length of the maximum match of the suffix string S i with the prefix string S j .

One way to program the overlap location is presented in the following listing.
Return for example. If we concatenate the strings S = {abc, cde, eab} with their overlaps, we get the string X = abcdeab with a length of 7, which is shorter than the previous one, but not the shortest. The shortest line is obtained when concatenating strings with overlaps in the order of 3-1-2, then the resulting string X = eabcde will have length 6. Thus, we reduced the problem to finding the optimal order of concatenated strings taking into account their overlaps.
Limitation All existing algorithms for finding the shortest superstructures assume that no string in the input set is a substring of any other string. Deletion of such lines can be made, for example, with the help of a suffix tree and, obviously, does not detract from the generality.
In this algorithm, at each iteration we try to find the maximum overlap of any two lines and merge them into one. We hope that the result of this algorithm will be close to the actual optimal one.
1. While S contains more than one line, we find two lines with the maximum overlap and merge them into one (for example, ABCD + CDEFG = ABCDEFG).
2. We return one line remaining in S.
An example of the implementation of this algorithm is presented in the following listing.
In this listing, another simple function, merge, appeared, which merges two lines into one, taking into account their overlap. Since the latter has already been computed, it is logical to simply pass it as a parameter.
One example ( worst case ):
● Input : S = {"ab k ", "b k c", "b k + 1 "}
● The output of the greedy algorithm: "ab k cb k + 1 " with a length of 4 + 4k
● Conclusion of the optimal algorithm: “ab k + 1 c” with a length of 4 + 2k
Worst case ratio
Generally speaking, this algorithm does not have a name and no one calls it as I do. It is called simply a 4-approximate algorithm for constructing the shortest superstructure. But since Blum, Young, Lee, Tromp and Yannakakis invented it, why not give this title?
For clarity , during the analysis of the algorithm, I will give an example of constructing an overlay for the set S = {S 0 : “cde”, S 1 : “abc”, S 2 : “eab”, S 3 : “fgh”, S 4 : “ ghf ”, S 5 :“ hed ”} .
The main idea of the algorithm is to find a covering with cycles of minimum full length for a complete directed weighted graph, the vertices of which are rows of a given set, and the weight of the edge from S i to S j is equal to overlap (S i , S j ) for all i, j.
The graph for our example (the vertices for visibility are signed i instead of S i ):

In my implementation, I will present this graph as an adjacency matrix , which will be formed in a banal way, shown in the following listing (in section 16.17 [1] D. Gasfield argues that the matrix can be formed more efficiently, but the method of forming this matrix does not affect consideration of this algorithm).
For our example, the adjacency matrix will look like this:

After the adjacency matrix is constructed, there is a need to find coverage with cycles of minimum full length. Such coverage can be found by reducing this task to the “assignment problem”. So, the task was to find the full purpose of the maximum weight (as it turned out, this step of the algorithm takes dominant time). Faster and easier ([1] p.16.19.13) this appointment can be found by the greedy method.
Greedy appointment ([1] p.16.19.13)
Baseline: k × k matrix A.
Result: full appointment M.
M can be represented in the code as an array (int [] M = new int [k]) and the first part of instruction 2.2 can be treated as M [i] = j; since in the full assignment each number from 0 to k occurs exactly once as the index of the row and exactly once as the index of the column.
An example of the implementation of the full assignment calculation is presented in the following listing.
Step-by-step calculation of the full assignment of the maximum weight by the greedy method for our example:

Now that the full assignment of the maximum weight is calculated, it is necessary to calculate the coverage with cycles of the minimum full length. For this you need:
taking the beginning of any (logical - zero) index of the full assignment, put it in a cycle, mark it used and see if its value equals the index of the beginning of the cycle? If yes, finish the cycle, put it in the cycle covering and start a new cycle, taking the first unlabeled element as the start. If not, take this value as an index and repeat the procedure until all items are marked.
The view of the cycle coverage in memory can be organized as a list of indexes of rows.
Algorithm listing (assign - full assignment):
For our example, finding the coverage in cycles will look like this:
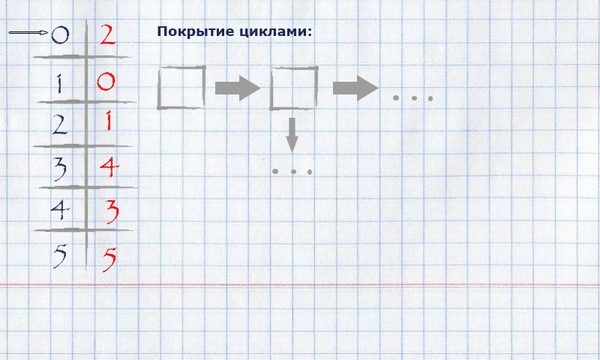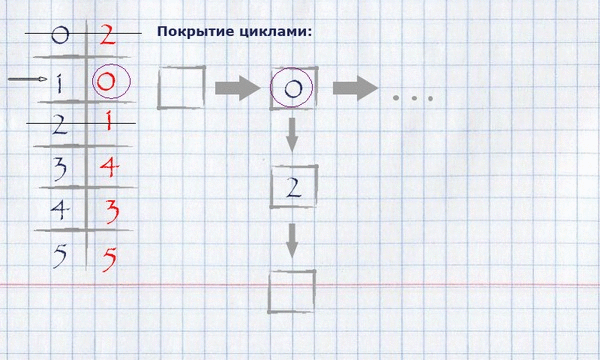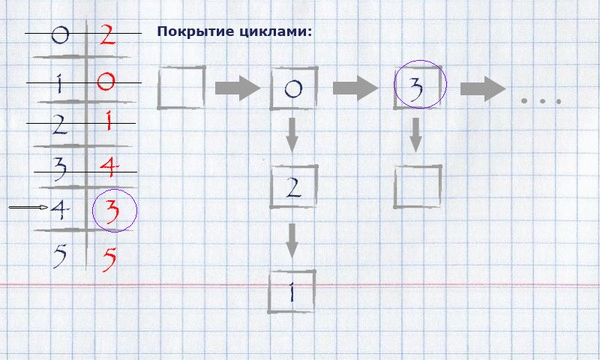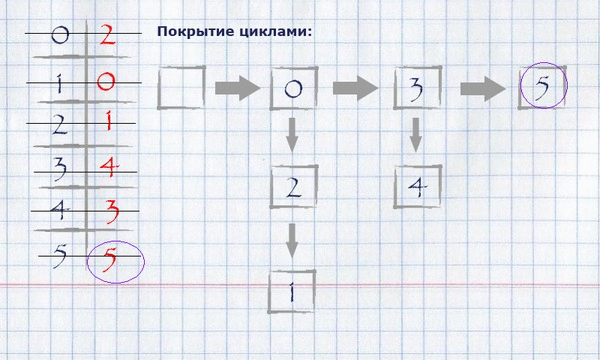
The resulting covering by cycles can be displayed on the initial graph, leaving only those edges that fall into the covering on it.

Assembly. Now that all the cycles in the coverage have been calculated, you can proceed to the assembly, directly, of the superline.
For this, we need the function prefix, which accepts the strings S 1 and S 2 and returns the string S 1 , truncated to the right of the overlap (S 1 , S 2 ) characters. But since all the overlaps have been calculated by us for a long time, the function can be written so that it accepts only one line and the number of characters it needs to be shortened by.
Now, for each cycle C i = {S 1 , S 2 , ..., S k-1 , S k }, it is necessary to make up the superscript X Ci = prefix (S 1 , overlap (S 1 , S 2 )) ++ prefix (S 2 , overlap (S 2 , S ...)) ++ prefix (S ..., overlap (S ..., S k-1 )) ++ S k .
The problem is that, since the cycles in the coverage are cycles , it means that we can cycle them and it is not clear from which line to start designing the X Ci superline . The answer is simple - you need to move the loop so as to minimize the overlap of the last and first line
Consider the first cycle of cyclic coverage from the example.

If you do not minimize the overlap of the last and first lines, but take C 0 = {1, 0, 2}, then the superscript is X C0 = abc ++ cde ++ eab = abcdeab, obviously, not the shortest possible one. The minimum overlap in this loop is 1, which means that any of the sequences {0, 2, 1} (cde ++ eab ++ abc = cdeabc) or {2, 1, 0} (eab ++ abc ++ cde = eabcde).
The following is the code that cyclically shifts each cycle in the coverage so as to minimize the overlaps of the first and last lines of each cycle, and constructs superscripts for each cycle.
Now we have the shortest sutures for each cycle in the coverage. The considered algorithm with factor 4 assumes a simple concatenation of these lines.
The source code for the greedy (Greedy.java) and Blum-Yang-Lee-Tromp-Yannakakis (Assign.java) algorithms are available in the git repository at bitbucket.org/andkorsh/scss/src . Also, there is the executable class Main.java, which when launched asks for the number of launches for measuring the speed of the algorithms and the path to the input file. Also, the Main class is able to generate input data itself, for this, instead of the file name, you must enter “random”, after which the number of lines, the maximum line length, the fixed length or not, and the number of characters in the alphabet will be requested. The report will be written to the file output.txt.
Source codes are guaranteed to be compiled by javac, starting with version “1.6.0_05”.
Testing is performed using the test function of the Main class.
[1] : Dan Gasfield. Strings, trees and sequences in algorithms. Computer science and computational biology. Nevsky Dialect, BHW-Petersburg, 2003. - 656 pp .: ISBN 5-7940-0103-8, 5-94157-321-9, 0-521-58519-8.
www.ozon.ru/context/detail/id/1393109
[2] : Dan Gusfield. Algorithms on Strings, Trees and Sequences: Computer Science and Computational Biology. THE PRESS SYNDICATE OF THE UNIVERSITY OF CAMBRIDGE, 1997 - 534 pp .: ISBN-10: 0521585198, ISBN-13: 978-0521585194.
www.amazon.com/Algorithms-Strings-Trees-Sequences-Computational/dp/0521585198
[3] : Shunji Li, Wenzheng Chi. Lecture # 3: Shortest Common Superstring - CS 352 (Advanced Algorithms), Spring 2011.
cs.carleton.edu/faculty/dlibenno/old/cs352-spring11/L03-shortest-superstring.pdf
When I searched the network for information on this issue and its solution in Russian, there were only a couple of posts about bioinformatics, where these words are mentioned in passing. The code (except for the greedy algorithm), of course, was not there either. Having understood the problem, this fact was inspired by the article here.
Be careful, 4 megabytes!
For the most part, the article is an attempt to translate into understandable language, illustrate, spice up with an example, and, of course, implement a 4-approximate algorithm for constructing superstructures from Dan Gasfield's book (see the used literature). But first, a small introduction and a 2-approximate, greedy algorithm.
')
In the simplest case (if the name of the problem did not include the word “shortest”), the solution of the problem would be a simple concatenation of the input strings in an arbitrary order. But in this formulation of the problem, we are dealing with NP-completeness, which means that at present there is no algorithm by which one could solve this problem on a Turing machine in a time that does not exceed the polynomial of the data size.
Input: S 1 , S 2 , ..., S n - the set of lines of a finite alphabet E *.
Output: X is the string of the alphabet E * containing each string S 1..n as a substring, where the length | X | minimized.
Example. Take the set of lines S = {abc, cde, eab}. In the case of concatenation, the length of the output superline will be 9 (X = abccdeeab), which is obviously not the best option, since the lines can have a suffix-prefix match. We will call the length of this coincidence overlap . (The choice of an English word is not accidental, since it does not have a specific and unambiguous translation into Russian. In the Russian-language literature, the terms overlay, intersection, overlap, and suffix-prefix coincidence are commonly used).
Retreat. The concept of overlap is one of the most important in the implementation process of the construction of superstructures. Calculating overlap for an ordered pair of strings (S i , S j ) is reduced to finding the length of the maximum match of the suffix string S i with the prefix string S j .

One way to program the overlap location is presented in the following listing.
/* * s1 * s2 ( s1 s2) */ private static int overlap(String s1, String s2) { int s1last = s1.length() - 1; int s2len = s2.length(); int overlap = 0; for (int i = s1last, j = 1; i > 0 && j < s2len; i--, j++) { String suff = s1.substring(i); String pref = s2.substring(0, j); if (suff.equals(pref)) overlap = j; } return overlap; } Return for example. If we concatenate the strings S = {abc, cde, eab} with their overlaps, we get the string X = abcdeab with a length of 7, which is shorter than the previous one, but not the shortest. The shortest line is obtained when concatenating strings with overlaps in the order of 3-1-2, then the resulting string X = eabcde will have length 6. Thus, we reduced the problem to finding the optimal order of concatenated strings taking into account their overlaps.
Limitation All existing algorithms for finding the shortest superstructures assume that no string in the input set is a substring of any other string. Deletion of such lines can be made, for example, with the help of a suffix tree and, obviously, does not detract from the generality.
Greedy algorithm
In this algorithm, at each iteration we try to find the maximum overlap of any two lines and merge them into one. We hope that the result of this algorithm will be close to the actual optimal one.
1. While S contains more than one line, we find two lines with the maximum overlap and merge them into one (for example, ABCD + CDEFG = ABCDEFG).
2. We return one line remaining in S.
An example of the implementation of this algorithm is presented in the following listing.
/* * , * ( ). */ public static String createSuperString(ArrayList<String> strings) { while (strings.size() > 1) { int maxoverlap = 0; String msi = strings.get(0), msj = strings.get(1); for (String si : strings) for (String sj : strings) { if (si.equals(sj)) continue; int curoverlap = overlap(si, sj); if (curoverlap > maxoverlap) { maxoverlap = curoverlap; msi = si; msj = sj; } } strings.add(merge(msi, msj, maxoverlap)); strings.remove(msi); strings.remove(msj); } return strings.get(0); } In this listing, another simple function, merge, appeared, which merges two lines into one, taking into account their overlap. Since the latter has already been computed, it is logical to simply pass it as a parameter.
/* * s1 s2 len, * overlap(s1, s2) */ private static String merge(String s1, String s2, int len) { s2 = s2.substring(len); return s1 + s2; } One example ( worst case ):
● Input : S = {"ab k ", "b k c", "b k + 1 "}
● The output of the greedy algorithm: "ab k cb k + 1 " with a length of 4 + 4k
● Conclusion of the optimal algorithm: “ab k + 1 c” with a length of 4 + 2k
Worst case ratio

Blum-Yang-Lee-Tromp-Yannakakis 4-approximation algorithm
Generally speaking, this algorithm does not have a name and no one calls it as I do. It is called simply a 4-approximate algorithm for constructing the shortest superstructure. But since Blum, Young, Lee, Tromp and Yannakakis invented it, why not give this title?
For clarity , during the analysis of the algorithm, I will give an example of constructing an overlay for the set S = {S 0 : “cde”, S 1 : “abc”, S 2 : “eab”, S 3 : “fgh”, S 4 : “ ghf ”, S 5 :“ hed ”} .
The main idea of the algorithm is to find a covering with cycles of minimum full length for a complete directed weighted graph, the vertices of which are rows of a given set, and the weight of the edge from S i to S j is equal to overlap (S i , S j ) for all i, j.
The graph for our example (the vertices for visibility are signed i instead of S i ):

In my implementation, I will present this graph as an adjacency matrix , which will be formed in a banal way, shown in the following listing (in section 16.17 [1] D. Gasfield argues that the matrix can be formed more efficiently, but the method of forming this matrix does not affect consideration of this algorithm).
int n = strings.size(); // overlap' // (Si, Sj). int[][] overlaps = new int[n][n]; for (int i = 0; i < n; i++) for (int j = 0; j < n; j++) overlaps[i][j] = overlap(strings.get(i), strings.get(j)); For our example, the adjacency matrix will look like this:

After the adjacency matrix is constructed, there is a need to find coverage with cycles of minimum full length. Such coverage can be found by reducing this task to the “assignment problem”. So, the task was to find the full purpose of the maximum weight (as it turned out, this step of the algorithm takes dominant time). Faster and easier ([1] p.16.19.13) this appointment can be found by the greedy method.
Greedy appointment ([1] p.16.19.13)
Baseline: k × k matrix A.
Result: full appointment M.
- Put M = Ø and declare all cells A available.
- while in A there are available cells do begin
- Among the available cells A, choose the cell (i, j) of the greatest weight.
- Place the cell (i, j) in M and make the cells in row i and column j inaccessible.
- end;
- Issue full M. appointment.
M can be represented in the code as an array (int [] M = new int [k]) and the first part of instruction 2.2 can be treated as M [i] = j; since in the full assignment each number from 0 to k occurs exactly once as the index of the row and exactly once as the index of the column.
An example of the implementation of the full assignment calculation is presented in the following listing.
/* * , * . - O(k*k*log(k)) */ private static int[] assignment(int[][] a) { int n = a.length; boolean[][] notallow = new boolean[n][n]; int[] m = new int[n]; while (true) { int max = -1, maxi = -1, maxj = -1; for (int i = 0; i < n; i++) { for (int j = 0; j < n; j++) { if (notallow[i][j]) continue; if (a[i][j] > max) { max = a[i][j]; maxi = i; maxj = j; } } } if (max == -1) break; m[maxi] = maxj; for (int i = 0; i < n; i++) { notallow[maxi][i] = true; notallow[i][maxj] = true; } } return m; } Step-by-step calculation of the full assignment of the maximum weight by the greedy method for our example:

Now that the full assignment of the maximum weight is calculated, it is necessary to calculate the coverage with cycles of the minimum full length. For this you need:
taking the beginning of any (logical - zero) index of the full assignment, put it in a cycle, mark it used and see if its value equals the index of the beginning of the cycle? If yes, finish the cycle, put it in the cycle covering and start a new cycle, taking the first unlabeled element as the start. If not, take this value as an index and repeat the procedure until all items are marked.
The view of the cycle coverage in memory can be organized as a list of indexes of rows.
Algorithm listing (assign - full assignment):
// // assign ArrayList<ArrayList<Integer>> cycles = new ArrayList<ArrayList<Integer>>(); ArrayList<Integer> cycle = new ArrayList<Integer>(); boolean[] mark = new boolean[assign.length]; for (int i = 0; i < assign.length; i++) { if (mark[i]) continue; cycle.add(i); mark[i] = true; if (assign[i] == cycle.get(0)) { cycles.add(cycle); cycle = new ArrayList<Integer>(); i = 0; } else { i = assign[i] - 1; } } For our example, finding the coverage in cycles will look like this:

The resulting covering by cycles can be displayed on the initial graph, leaving only those edges that fall into the covering on it.

Assembly. Now that all the cycles in the coverage have been calculated, you can proceed to the assembly, directly, of the superline.
For this, we need the function prefix, which accepts the strings S 1 and S 2 and returns the string S 1 , truncated to the right of the overlap (S 1 , S 2 ) characters. But since all the overlaps have been calculated by us for a long time, the function can be written so that it accepts only one line and the number of characters it needs to be shortened by.
/* * s1, * ov */ private static String prefix(String s1, int ov) { return s1.substring(0, s1.length() - ov); } Now, for each cycle C i = {S 1 , S 2 , ..., S k-1 , S k }, it is necessary to make up the superscript X Ci = prefix (S 1 , overlap (S 1 , S 2 )) ++ prefix (S 2 , overlap (S 2 , S ...)) ++ prefix (S ..., overlap (S ..., S k-1 )) ++ S k .
The problem is that, since the cycles in the coverage are cycles , it means that we can cycle them and it is not clear from which line to start designing the X Ci superline . The answer is simple - you need to move the loop so as to minimize the overlap of the last and first line
Consider the first cycle of cyclic coverage from the example.

If you do not minimize the overlap of the last and first lines, but take C 0 = {1, 0, 2}, then the superscript is X C0 = abc ++ cde ++ eab = abcdeab, obviously, not the shortest possible one. The minimum overlap in this loop is 1, which means that any of the sequences {0, 2, 1} (cde ++ eab ++ abc = cdeabc) or {2, 1, 0} (eab ++ abc ++ cde = eabcde).
The following is the code that cyclically shifts each cycle in the coverage so as to minimize the overlaps of the first and last lines of each cycle, and constructs superscripts for each cycle.
// , // overlap // . ArrayList<String> superstrings = new ArrayList<String>(); for (ArrayList<Integer> c : cycles) { String str = ""; ArrayList<Integer> ovs = new ArrayList<Integer>(); for (int i = 0; i < c.size() - 1; i++) ovs.add(overlaps[c.get(i)][c.get(i+1)]); int min = overlaps[c.get(c.size()-1)][c.get(0)]; int shift = 0; for (int i = 0; i < ovs.size(); i++) if (ovs.get(i) < min) { min = ovs.get(i); shift = i + 1; } Collections.rotate(c, -shift); for (int i = 0; i < c.size() - 1; i++) str += prefix(strings.get(c.get(i)), overlaps[c.get(i)][c.get(i+1)]); str += strings.get(c.get(c.size()-1)); superstrings.add(str); } Now we have the shortest sutures for each cycle in the coverage. The considered algorithm with factor 4 assumes a simple concatenation of these lines.
// superstrings StringBuilder superstring = new StringBuilder(); for (String str : superstrings) superstring.append(str); return superstring.toString(); Source codes and testing
The source code for the greedy (Greedy.java) and Blum-Yang-Lee-Tromp-Yannakakis (Assign.java) algorithms are available in the git repository at bitbucket.org/andkorsh/scss/src . Also, there is the executable class Main.java, which when launched asks for the number of launches for measuring the speed of the algorithms and the path to the input file. Also, the Main class is able to generate input data itself, for this, instead of the file name, you must enter “random”, after which the number of lines, the maximum line length, the fixed length or not, and the number of characters in the alphabet will be requested. The report will be written to the file output.txt.
Source codes are guaranteed to be compiled by javac, starting with version “1.6.0_05”.
Testing is performed using the test function of the Main class.
private static int test(ArrayList<String> substrings, String superstring) { int errors = 0; for (String substring : substrings) if (superstring.indexOf(substring) < 0) errors++; return errors; } References
[1] : Dan Gasfield. Strings, trees and sequences in algorithms. Computer science and computational biology. Nevsky Dialect, BHW-Petersburg, 2003. - 656 pp .: ISBN 5-7940-0103-8, 5-94157-321-9, 0-521-58519-8.
www.ozon.ru/context/detail/id/1393109
[2] : Dan Gusfield. Algorithms on Strings, Trees and Sequences: Computer Science and Computational Biology. THE PRESS SYNDICATE OF THE UNIVERSITY OF CAMBRIDGE, 1997 - 534 pp .: ISBN-10: 0521585198, ISBN-13: 978-0521585194.
www.amazon.com/Algorithms-Strings-Trees-Sequences-Computational/dp/0521585198
[3] : Shunji Li, Wenzheng Chi. Lecture # 3: Shortest Common Superstring - CS 352 (Advanced Algorithms), Spring 2011.
cs.carleton.edu/faculty/dlibenno/old/cs352-spring11/L03-shortest-superstring.pdf
Source: https://habr.com/ru/post/201656/
All Articles