Simulate the electrical activity of neurons
Introduction
Immediately I will inform you that this article is not related to perceptrons, Hopfield networks or any other artificial neural networks. We will model the work of a “real”, “living”, biological neural network in which the processes of generation and propagation of nerve impulses take place. In English-language literature, such networks are called spiking neural networks due to their difference from artificial neural networks, while Russian-language literature does not have a well-established name. Someone calls them just neural networks, someone - impulse neural networks, and someone - spike.
Probably, the majority of readers have heard about the Blue Brain and Human Brain projects sponsored by the European Union. The EU government issued about a billion euros for the last project, indicating that there is a lot of interest in this area. Both projects are closely related and intersect with each other, even the head of them is common, Henry Markram , which can create some confusion about how they differ from each other. In short, the ultimate goal of both projects is to develop a model of the whole brain, all ~ 86 billion neurons. The Blue Brain Project is the computational part, and the Human Brain is the more fundamental part, where they are working on collecting scientific data on the principles of the brain and creating a single model. To touch this science and try to do something similar, although on a much smaller scale, this note was written.
On Habré there were already several interesting and informative articles on neurobiology, which is very pleasing.
1. Neurobiology and artificial intelligence: part one - educational program.
2. Neurobiology and artificial intelligence: the second part is the intellect and the presentation of information in the brain.
3. Neurobiology and artificial intelligence: part three - data presentation and memory
But they did not address issues of computational neuroscience, or otherwise computational neuroscience, which includes computer modeling of the electrical activity of neurons, so I decided to fill this gap.
')
Some biology
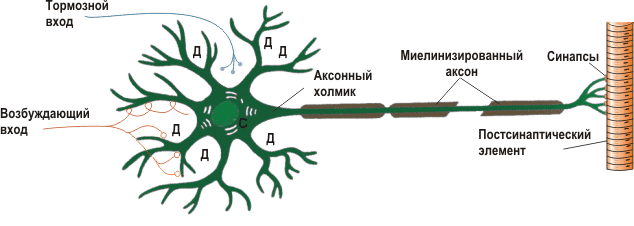
Fig. 1 - Schematic representation of the structure of the neuron.
Before we start modeling, we need to familiarize ourselves with some of the basics of neurobiology. A typical neuron consists of 3 parts: the body (soma), dendrites and axon. Dendrites receive a signal from other neurons (this is the input of a neuron), and an axon transmits signals from the body of a neuron to other neurons (output). The site of contact of the axon of one neuron and the dendrite of another neuron is called a synapse. The signal received from the dendrites is summed up in the body and if it exceeds a certain threshold, a nerve impulse is generated or a spike in a different way. The cell body is surrounded by a lipid membrane, which is a good insulator. The ionic composition of the cytoplasm of the neuron and the extracellular fluid differ. In the cytoplasm, the concentration of potassium ions is higher, and the concentration of sodium and chlorine is lower; in the intercellular fluid, the opposite is true. This is due to the work of ionic pumps, which constantly pump over certain types of ions against the concentration gradient, while consuming the energy stored in Adenosine Tri Phosphate (ATP) molecules. The most famous and studied of these pumps is the sodium-potassium pump. It brings 3 sodium ions to the outside, and inside the neuron takes 2 potassium ions. Figure 2 shows the ionic composition of the neuron and marked ion pumps. Due to the operation of these pumps, an equilibrium potential difference between the inner side of the membrane, which is negatively charged, and the external one that is positively is formed in the neuron.

Fig. 2 - Ion composition of the neuron and the environment
In addition to the pumps, there are ion channels on the surface of the neuron, which, when the potential changes or under chemical exposure, can open or close, thereby increasing or decreasing the currents of a certain type of ion. If the membrane potential exceeds a certain threshold, sodium channels open, and since there is more sodium outside, an electric current directed inside the neuron arises, which further increases the membrane potential and opens the sodium channels even more, a sharp increase in the membrane potential occurs. Physicists will call this positive feedback. But, starting from some potential value higher than the threshold potential of opening sodium channels, potassium channels are also opened, due to which potassium ions begin to flow outwards, reducing the membrane potential and thereby returning it to an equilibrium value. If the initial excitation is less than the threshold for opening sodium channels, then the neuron will return to its equilibrium state. Interestingly, the amplitude of the generated pulse weakly depends on the amplitude of the exciting current: either there is a pulse, or there is none, the “all or nothing” law.
By the way, it was the “all or nothing” principle that inspired McCulloch and Pitts to create models of artificial neural networks. But the field of artificial neural networks is developing in its own way, and its main goal is the most optimal solution of practical problems, irrespective of how this relates to the processing of information in the living brain. While spike neural networks are a model of the operation of a real brain. It is possible to assemble a spike network for recognizing visual images, but classical neural networks are better suited for practical use, they are simpler, are considered to be faster on a computer, and many algorithms are invented for training for specific practical tasks.
The principle of “all or nothing” is clearly illustrated in Figure 3. The input current directed to the inner side of the neuron membrane is shown below, and the potential difference between the inner and outer side of the membrane is above. Therefore, according to the currently dominant concept in living neural networks, information is encoded in the times when pulses occur, or, as physicists would say, by phase modulation.
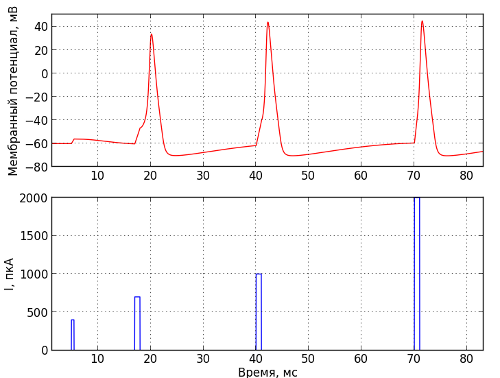
Fig. 3 - The generation of nerve impulses. The bottom shows the current supplied to the cell in the PCA, and at the top is the membrane potential in mV
It is possible to excite a neuron, for example, by thrusting a microelectrode into it and applying current to the neuron, but in the living brain, the excitation usually takes place by means of a synaptic effect. As already mentioned, the neurons are connected to each other using synapses, which are formed at the contact points of the axon of one neuron with the dendrites of the other. The neuron from which the signal is coming is called presynaptic, and the one to which the signal goes is called postsynaptic. When a pulse arises on a presynaptic neuron, it will emit neurotransmitters into the synaptic cleft, which open sodium channels on the postsynaptic neuron, and then a chain of the above-described events occurs that lead to excitation. In addition to excitation, neurons can also inhibit each other. If the presynaptic neuron is inhibitory, it will release the inhibitory neurotransmitter opening the chlorine channels into the synaptic cleft, and since there is more chlorine outside, chlorine flows into the neuron because of which the negative charge on the inner side of the membrane increases (do not forget that chlorine ions in differences from sodium and potassium are negatively charged), driving the neuron to an even more inactive state. In this state, the neuron is more difficult to excite.
Mathematical model of neuron
Based on the dynamic mechanisms of the neuron operation described above, its mathematical model can be compiled. At present, various relatively simple models have been created, such as “Inregrate and Fire”, in which the neuron is represented as a capacitor and a resistor, as well as more complex, biologically plausible, models, such as the Hodgkin-Huxley model, which is much more complicated as in computational terms so in terms of analyzing its dynamics, but it describes the dynamics of the membrane potential of a neuron much more accurately. In this article we will use the Izhikevich model [1], it is a compromise between computational complexity and biophysical plausibility. Despite its computational simplicity, this model can reproduce a large number of phenomena occurring in real neurons. The Izhikevich model is defined as a system of differential equations (Figure 4).

Fig. 4 - Izhikevich model
Where a, b, c, d, k, Cm are different parameters of the neuron. Vm is the potential difference on the inside and outside of the membrane, and Um is an auxiliary variable. I is the external direct current applied. In this model, such neuron-specific properties are observed as: spike generation in response to a single external current pulse and generation of a spike sequence with a certain frequency when a constant external current is applied to the neuron. Isyn is the sum of synaptic currents from all neurons with which this neuron is connected.
If a spike is generated on a presynaptic neuron, a synaptic current jump occurs on the postsynaptic neuron, which exponentially decays with a characteristic time.
Go to coding
So, we proceed to the most interesting. It's time to code a virtual piece of nervous tissue on a computer. To do this, we will numerically solve a system of differential equations that define the dynamics of the membrane potential of a neuron. For the integration we will use the Euler method. We will code in C ++, draw using scripts written in Python using the Matplolib library, but those who do not have Python can draw using Exel.
We will need two-dimensional arrays Vms, Ums of Tsim * Nneur dimension for storing membrane potentials and auxiliary variables of each neuron, at each moment in time, Tsim is the simulation time in readings, and Nneur is the number of neurons in the network.
Links will be stored in the form of two arrays of pre_con and post_con of the dimension Ncon , where the indices are the numbers of the links, and the values are the indices of presynaptic and postsynaptic neurons. Ncon is the number of connections.
We also need an array to represent a variable modulating the exponentially decaying postsynaptic current of each synapse, for this we create an array y of the dimension Ncon * Tsim .
const float h = .5f; // const int Tsim = 1000/.5f; // const int Nexc = 100; // (excitatory) const int Ninh = 25; // (inhibitory) const int Nneur = Nexc + Ninh; const int Ncon = Nneur*Nneur*0.1f; // , 0.1 2- float Vms[Nneur][Tsim]; // float Ums[Nneur][Tsim]; // float Iex[Nneur]; // float Isyn[Nneur]; // int pre_conns[Ncon]; // int post_conns[Ncon]; // float weights[Ncon]; // float y[Ncon][Tsim]; // float psc_excxpire_time = 4.0f; // , float minWeight = 50.0f; // , float maxWeight = 100.0f; // float Iex_max = 40.0f; // 50 float a = 0.02f; float b = 0.5f; float c = -40.0f; // float d = 100.0f; float k = 0.5f; float Vr = -60.0f; float Vt = -45.0f; float Vpeak = 35.0f; // , float V0 = -60.0f; // float U0 = 0.0f; // float Cm = 50.0f; // , As already mentioned, the information is encoded in the time of occurrence of pulses, so we create arrays to save the time of their occurrence and the indices of neurons where they originated. Then they can be written to a file for visualization purposes.
float spike_times[Nneur*Tsim]; // int spike_neurons[Nneur*Tsim]; // int spike_num = 0; // We scatter randomly links and set weights.
void init_connections(){ for (int con_idx = 0; con_idx < Ncon; ){ // pre_conns[con_idx] = rand() % Nneur; post_conns[con_idx] = rand() % Nneur; weights[con_idx] = (rand() % ((int)(maxWeight - minWeight)*10))/10.0f + minWeight; if (pre_conns[con_idx] >= Nexc){ // weights[con_idx] = -weights[con_idx]; } con_idx++; } } Setting the initial conditions for neurons and the random setting of the external applied current. Those neurons for which the external current exceeds the spike generation threshold will generate spikes with a constant frequency.
void init_neurons(){ for (int neur_idx = 0; neur_idx < Nneur; neur_idx++){ // Iex[neur_idx] = (rand() % (int) (Iex_max*10))/10.0f; Isyn[neur_idx] = 0.0f; Vms[neur_idx][0] = V0; Ums[neur_idx][0] = U0; } } The main part of the program with the integration of the Izhikevich model.
float izhik_Vm(int neuron, int time){ return (k*(Vms[neuron][time] - Vr)*(Vms[neuron][time] - Vt) - Ums[neuron][time] + Iex[neuron] + Isyn[neuron])/Cm; } float izhik_Um(int neuron, int time){ return a*(b*(Vms[neuron][time] - Vr) - Ums[neuron][time]); } int main(){ init_connections(); init_neurons(); float expire_coeff = exp(-h/psc_excxpire_time); // for (int t = 1; t < Tsim; t++){ // for (int neur = 0; neur < Nneur; neur++){ Vms[neur][t] = Vms[neur][t-1] + h*izhik_Vm(neur, t-1); Ums[neur][t] = Ums[neur][t-1] + h*izhik_Um(neur, t-1); Isyn[neur] = 0.0f; if (Vms[neur][t-1] > Vpeak){ Vms[neur][t] = c; Ums[neur][t] = Ums[neur][t-1] + d; spike_times[spike_num] = t*h; spike_neurons[spike_num] = neur; spike_num++; } } // for (int con = 0; con < Ncon; con++){ y[con][t] = y[con][t-1]*expire_coeff; if (Vms[pre_conns[con]][t-1] > Vpeak){ y[con][t] = 1.0f; } Isyn[post_conns[con]] += y[con][t]*weights[con]; } } save2file(); return 0; } The full text of the code can be downloaded here . The code is freely compiled and launched even under Windows with Visual Studio 2010 Express Edition or MinGW, even under GNU / Linux with GCC. Upon completion, the program saves the raster activity, times and indices of the appearance of nerve impulses, and oscillograms of the average membrane potential in the rastr.csv and oscill.csv files, respectively. You can directly them in Exel and draw. Or with the help of the Python scripts attached by me, but for their work the Matplotlib library is needed. For those with GNU / Linux, installing the python-matplotlib package from the repositories is easy, while Windows users will have to manually download the numpy, scypy, pyparsing, python-dateutil and matplotlib packages from here.
Drawing an activity raster - plot_rastr.py
Drawing the average membrane potential - plot_oscill.py
results

Fig. 5 - Neural network activity. At the top along the y-axis, the numbers of neurons are plotted, and the times when the spike is generated on the neuron are marked by a dot, below is the average number of spikes per 1 ms of time.

Fig. 6 - Average membrane potential, “electroencephalogram”.
This is what is obtained by simulating 1 second of activity of 125 neurons. We observe periodic activity at a frequency of ~ 3 Hz, similar to a delta rhythm .
[1] EM Izhikevich, Dynamic Systems in Neuroscience: The Geometry of Excitability and Bursting, USA, MA, Cambridge: The MIT Press., 2007
Source: https://habr.com/ru/post/201220/
All Articles