Algorithms in bioinformatics, online course
 Sequencing of the human genome ten years ago was the cause of the computational revolution in biology. It has become an incentive to create more new algorithms than in any other fundamental area of science.
Sequencing of the human genome ten years ago was the cause of the computational revolution in biology. It has become an incentive to create more new algorithms than in any other fundamental area of science.On October 21, Phillip Compo and Pavel Pevzner from the University of California and I launch an online course on bioinformatics algorithms on Coursera. Already before October 21, you can see the content of the first chapter of the course and solve the problems on our new educational project Stepic, which is being worked on by the team of the Rosalind project, which is well-known in narrow (bioinformatics) circles.
Genome sequencing is only one of hundreds of biological problems that have become inextricably linked with the computational methods necessary to solve them. This course will cover algorithmic ideas that are fundamental to understanding modern biology. Computational concepts such as dynamic programming and graph theory will help study algorithms used in a wide range of biological tasks: assembling genomes, comparing DNA sequences and proteins, searching for regulatory motives, analyzing genome rearrangements, identifying proteins, restoring the tree of life, and many others. Throughout the course, participants will apply real bioinformatics algorithms to real genetic data.
')
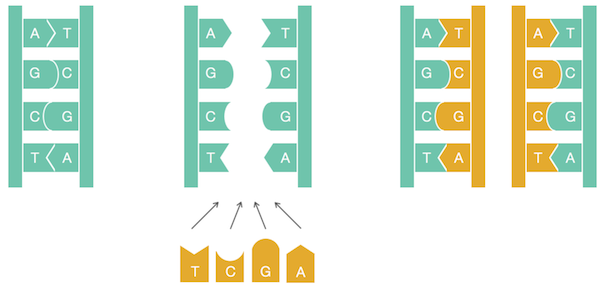
Each chapter of the course contains a separate biological topic. For example, the first chapter is devoted to finding the starting points of replication (positions in the genome from which DNA begins to divide in the cell), and how this, at first glance, purely biological problem can be solved with the help of simple programming and analysis of genomic sequences.
Weekly course program
- Find replication start points: Where Does DNA Replication Begin? (Algorithmic Warm-up)
- Computational Mass Spectrometry: How Do We Sequence Antibiotics? (Brute Force Algorithms)
- Search for frequently repeated sequences (regulatory motifs): Which DNA Patterns Act As Cellular Clocks? (Greedy and Randomized Algorithms)
- Genome Assembly: How Do We Assemble Genomes? (Graph Algorithms)
- Genomic restructuring: Are There Fragile Regions in the Human Genome? (Combinatorial Algorithms)
- Line alignment: How Do We Compare Biological Sequences? (Dynamic Programming Algorithms)
- Quick search for substrings, indexing strings: How Do We Locate Disease-Causing Mutations? (Combinatorial Pattern Matching)
Introductory video of Professor Pevzner
Massive Open Online Research
An important component of the course are open scientific tasks formulated for each chapter. The discussion on each task will be supervised by the leading bioinformatics scientist in each specific field. For example, open tasks from the first chapter of the course are supervised by Mikhail Gelfand (IITP RAS, MGU), Uri Keich (University of Sydney) and Glenn Tesler (UCSD).
Related Links
- Coursera: Bioinformatics Algorithms (Part 1)
- Stepic: Bioinformatics Algorithms: An Active-Learning Approach
- Course press release from University of California, San Diego
- Press release from the California Institute for Telecommunications and Information Technology
- UC San Diego Team To Intro Massive Open Online Research
- Preliminary detailed course program (pdf)
- Rosalind - a platform for studying bioinformatics
- Institute of Bioinformatics
Source: https://habr.com/ru/post/196870/
All Articles