Systematics of prokaryotes - detailed explanations
In the previous article I didn’t bother myself with a detailed description of the idea. It seemed to me to be intuitive, understandable and elementary. But after discussing with Davidov I realized that the idea was not so easy to grab. The fact is that now classical concepts in phylogenetics are built on a single dogma, which distorts the outlook of biologists.
When we build an evolutionary tree, we certainly want to know in what sequence in time the species have speculated. But classical phylogenetics announced that it is not a scientific question to ask. And essentially signed her helplessness. Indeed, it is difficult to judge the time of speciation, while the evolutionary process goes on every minute. But you can. Explain how this detailed explanation can and is intended.
')
Considering on the fingers, it will not be speculative speculative, so we consider the problem in one particular example.

Be sure to look at an example in high resolution .
Let's start with the top of Cellvibrio. Cellvibrio is one kind of proteobacteria. In the genus there are several species. But it is not the species that is sequenced, but one of the representatives of the species - the strain is called (it’s the same as taking the genome of a specific person - this is how we get the strain of the species). Now, in this example, it turned out that now only one Cellvibrio strain, the Cellvibrio japonicus species, is sequenced, and the stamp was named Ueda107. Therefore, on the graph at the top of Cellvibrio there is no connection with itself - there is no information.
Next, look at the other top Cellulomonas. This is a different genus, and two strains of different species are sequenced. And near the top there is a connection with itself. And there are numbers 8-8. This means that when comparing these strains (in this case only two) of this kind, it turned out that they have 100 different tRNAs that match 100%.
In general, the body usually has about 20 tRNA (for each amino acid). Further, as a rule, the body contains 2-3 variations of the same tRNA. But more ancient organisms do not even contain all 20 tRNAs, and often in one variant. Younger have greater diversity. Therefore, if within the genus if 40-60 tRNA coincide, then it is safe to say that this is a relatively young genus, and the strains are related to it correctly. In this case, we have 8 - this is not enough, but this is just an exception, because This is one of the oldest organisms.
Where does this come from (except from a small number of coincident tRNAs)?
In the last article I gave a complete graph of systematics of prokaryotes
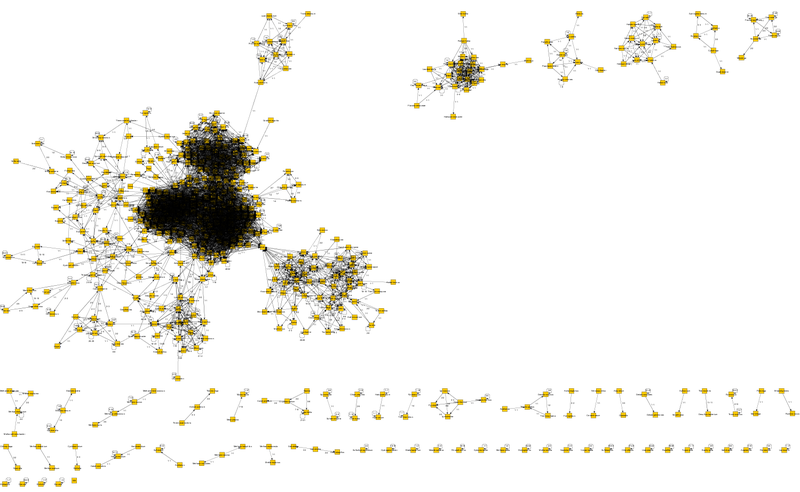
There is an ideal point between the biggest black spot (there are representatives of Gamma proteobacteria) and the area on the right, where individual nodes are visible. This point is the top of Cellvibrio.
What is it remarkable? The genus Cellvibrio itself belongs to gamma-proteobacteria, but we see that this genus has connections not only with a cluster of gamma-proteobacteria, but also with another, for example, with Cellulomonas apex - and this genus is classified as Actinobacteria.
The question arises: what is more likely that representatives of the genus Cellvibrio aggregated tRNA from a dozen gammaproteobacteria and a dozen other types of different types, for example from Actinobacteria?
(Separately, I emphasize that even the coincidence of one tRNA is not a random event, and here 5-7 coincides.)
Or should we conclude that the Cellvibrio japonicus species existed before proteobacteria and a number of types were formed, such as Actinobacteria?
Naturally, the probability of having one species is initially greater than the aggregation of several dozen species into one species.
Yes, at the present time Cellvibrio japonicus may differ from Cellvibrio japonicus which was a million years ago, but over this million years its 5-7 tRNA did not change at all, while proteins and, say, rRNA underwent mutations.
This is one of the simplest and most vivid examples of how to judge the age of species / genera in relation to others.
But there are very few such ideal points, as we see in the general graph, it is rather an accident when it is so easy to judge which species is older and which is younger.
More often, the layering of some genera on others, and it becomes more difficult to understand. Let's trace it by the first column. The Cellvibrio vertex has connections to all other vertices, with the exception of only one Nocardia, which is connected in the same way Cellvibrio - Cellulomonas - Nocardia. Those. we can say here that the genus Nocardia is younger than all the others, because requires the formation of an intermediate genus Cellulomonas.
How to analyze the rest of the relationship?
There are several ways, depending on the desire details. If you look at the signatures of the links there, we see the minimum-maximum number of identical tRNAs. For example, between the Cellvibrio and Isoptericola genera (located on the far left) there are 6 identical tRNAs. On the other hand, between Cellvibrio and Kocuria (rightmost), there is only 1 identical tRNA.
Thus, it can be said that the genera are removed from the Cellvibrio gradually from left to right.
This is true for the first row at 100%. And again, this allows us to talk about the time of birth of the childbirth.
But with the second row more difficult. The fact is that in the second row the tops have two entrances from Cellvibrio and Cellulomonas, and it becomes not clear from which of these two the next genus was formed.
You can do statistically. For example, let's compare two Cellvibrio links - Isoptericola and Cellulomonas - Isoptericola. The first has 6-6 identical tRNAs, the second 5-7. So we can conclude that 6 identical tRNAs were transferred from Cellvibrio to Cellulomonas, and only then the genus Isoptericola was formed to which another 1 tRNA was transferred, and in some form 1 tRNA was lost (5-7 total). And then the Cellvibrio-Isoptericola connection is mediated and can be removed. And it becomes more clearly visible who comes from whom in time.
There are other options. There it is already possible to look not statistically, but in detail, whether there are specific tRNAs as a prerequisite for the subsequent formation of a genus. I will explain in more detail on the same example in the comments - if there is interest.
Those. The basic idea is that the Whole cannot be formed if there are still no parts in nature of which the whole should consist.
When we build an evolutionary tree, we certainly want to know in what sequence in time the species have speculated. But classical phylogenetics announced that it is not a scientific question to ask. And essentially signed her helplessness. Indeed, it is difficult to judge the time of speciation, while the evolutionary process goes on every minute. But you can. Explain how this detailed explanation can and is intended.
')
Ideal point of divergence
Considering on the fingers, it will not be speculative speculative, so we consider the problem in one particular example.

Be sure to look at an example in high resolution .
Let's start with the top of Cellvibrio. Cellvibrio is one kind of proteobacteria. In the genus there are several species. But it is not the species that is sequenced, but one of the representatives of the species - the strain is called (it’s the same as taking the genome of a specific person - this is how we get the strain of the species). Now, in this example, it turned out that now only one Cellvibrio strain, the Cellvibrio japonicus species, is sequenced, and the stamp was named Ueda107. Therefore, on the graph at the top of Cellvibrio there is no connection with itself - there is no information.
Next, look at the other top Cellulomonas. This is a different genus, and two strains of different species are sequenced. And near the top there is a connection with itself. And there are numbers 8-8. This means that when comparing these strains (in this case only two) of this kind, it turned out that they have 100 different tRNAs that match 100%.
In general, the body usually has about 20 tRNA (for each amino acid). Further, as a rule, the body contains 2-3 variations of the same tRNA. But more ancient organisms do not even contain all 20 tRNAs, and often in one variant. Younger have greater diversity. Therefore, if within the genus if 40-60 tRNA coincide, then it is safe to say that this is a relatively young genus, and the strains are related to it correctly. In this case, we have 8 - this is not enough, but this is just an exception, because This is one of the oldest organisms.
Where does this come from (except from a small number of coincident tRNAs)?
In the last article I gave a complete graph of systematics of prokaryotes

There is an ideal point between the biggest black spot (there are representatives of Gamma proteobacteria) and the area on the right, where individual nodes are visible. This point is the top of Cellvibrio.
What is it remarkable? The genus Cellvibrio itself belongs to gamma-proteobacteria, but we see that this genus has connections not only with a cluster of gamma-proteobacteria, but also with another, for example, with Cellulomonas apex - and this genus is classified as Actinobacteria.
The question arises: what is more likely that representatives of the genus Cellvibrio aggregated tRNA from a dozen gammaproteobacteria and a dozen other types of different types, for example from Actinobacteria?
(Separately, I emphasize that even the coincidence of one tRNA is not a random event, and here 5-7 coincides.)
Or should we conclude that the Cellvibrio japonicus species existed before proteobacteria and a number of types were formed, such as Actinobacteria?
Naturally, the probability of having one species is initially greater than the aggregation of several dozen species into one species.
Yes, at the present time Cellvibrio japonicus may differ from Cellvibrio japonicus which was a million years ago, but over this million years its 5-7 tRNA did not change at all, while proteins and, say, rRNA underwent mutations.
This is one of the simplest and most vivid examples of how to judge the age of species / genera in relation to others.
Ordering by age
But there are very few such ideal points, as we see in the general graph, it is rather an accident when it is so easy to judge which species is older and which is younger.
More often, the layering of some genera on others, and it becomes more difficult to understand. Let's trace it by the first column. The Cellvibrio vertex has connections to all other vertices, with the exception of only one Nocardia, which is connected in the same way Cellvibrio - Cellulomonas - Nocardia. Those. we can say here that the genus Nocardia is younger than all the others, because requires the formation of an intermediate genus Cellulomonas.
How to analyze the rest of the relationship?
There are several ways, depending on the desire details. If you look at the signatures of the links there, we see the minimum-maximum number of identical tRNAs. For example, between the Cellvibrio and Isoptericola genera (located on the far left) there are 6 identical tRNAs. On the other hand, between Cellvibrio and Kocuria (rightmost), there is only 1 identical tRNA.
Thus, it can be said that the genera are removed from the Cellvibrio gradually from left to right.
This is true for the first row at 100%. And again, this allows us to talk about the time of birth of the childbirth.
But with the second row more difficult. The fact is that in the second row the tops have two entrances from Cellvibrio and Cellulomonas, and it becomes not clear from which of these two the next genus was formed.
You can do statistically. For example, let's compare two Cellvibrio links - Isoptericola and Cellulomonas - Isoptericola. The first has 6-6 identical tRNAs, the second 5-7. So we can conclude that 6 identical tRNAs were transferred from Cellvibrio to Cellulomonas, and only then the genus Isoptericola was formed to which another 1 tRNA was transferred, and in some form 1 tRNA was lost (5-7 total). And then the Cellvibrio-Isoptericola connection is mediated and can be removed. And it becomes more clearly visible who comes from whom in time.
There are other options. There it is already possible to look not statistically, but in detail, whether there are specific tRNAs as a prerequisite for the subsequent formation of a genus. I will explain in more detail on the same example in the comments - if there is interest.
Those. The basic idea is that the Whole cannot be formed if there are still no parts in nature of which the whole should consist.
Source: https://habr.com/ru/post/162797/
All Articles