Implementing C # principal component method
Hello. This week, in the course on machine learning, Professor Andrew Ng told the audience about the main component method with which you can reduce the dimension of the feature space of your data. But unfortunately, he did not talk about the method of calculating the eigenvectors and eigenvalues of the matrix, he simply said that it was difficult and advised using the matlab / Octave function [USV] = svd (a).
For my project, I needed the implementation of this method in c #, which I did today. The method of the main components is very elegant and beautiful, and if you do not understand the mathematics that lies behind all this, then it can all be called shamanism. The problem of calculating the eigenvectors of the matrix is that there is no quick way to calculate their exact values, so you have to get out. I want to talk about one of these ways to get out, as well as the c # code that performs this procedure. I ask under the cat.
')
Part one: algorithm initialization. The input to the algorithm is an array of data, as well as the dimension of the space to which it is necessary to reduce the data.
Part Two: Reducing the dimension of the input vector. At the input is a vector, dimensions as in the data array used in the initialization step; at the output, a vector of smaller dimension (or the projection of the input vector on an orthonormal basis formed by sampling from eigenvectors).
Part three: recovery of the dimension of the vector (of course, with the loss of information). The input is a vector of dimension equal to that to which we reduced the vectors; output vector of the original dimension.
As you can see, the entire computational complexity is in the initialization step when calculating the covariance matrix and eigenvectors. I will not stop on the calculation of the covariance matrix, since it is calculated naively by definition. Having studied several algorithms for calculating the eigenvectors of the matrix, I stopped at the iterative QR decomposition method. He has a significant limitation, he gives a less accurate result only for symmetric matrices, but we were lucky :), the covariance matrix is just that. The second limitation of this algorithm is that the matrix must be full-rank .
The iterative QR method for finding eigenvectors obviously uses QR decomposition , so first we have to implement this process. Gram-Schmidt algorithm was chosen to implement this process.
For this we need a function that calculates the projection of the vector a on the vector b :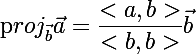 where <> - denotes the scalar product of vectors.
where <> - denotes the scalar product of vectors.
I will not dwell on even simpler functions, such as ScalarVectorProduct, so that the code size in the text does not exceed the critical limit.
So the actual QR decomposition procedure itself IList <double []> DecompositionGS (double [] [] a) receives a matrix as input, and returns two matrices in response:
First, we divide the matrix into columns and write them into the av list:
We initialize two auxiliary lists:
These are the same lists used in the algorithm scheme:

After initialization, the first u and e have been calculated, we continue in the loop to calculate the following values:
Finally, we generate data in the output format:
Hooray! Now you need a test for this:
The Helper.AreEqualMatrices function compares element-wise matrices with the accuracy of the third parameter.
The iterative QR method is based on the remarkable theorem, the essence of which is that if you initialize the matrix A zero as and repeat the following process infinitely many times:
and repeat the following process infinitely many times:
and then multiply all obtained Q , then the result will be a matrix, in the columns of which will be the eigenvectors of the original matrix, the values of which will be the more accurate, the longer the process was performed. In other words, when the number of iterations tends to infinity, the product will tend to the exact values of the eigenvectors. At the same time last
will tend to the exact values of the eigenvectors. At the same time last  will on the main diagonal contain eigenvalues of the matrix, approximate of course. I remind you that this algorithm works more or less precisely only for symmetric matrices.
will on the main diagonal contain eigenvalues of the matrix, approximate of course. I remind you that this algorithm works more or less precisely only for symmetric matrices.
So, the QR iteration method IList <double [] []> EigenVectorValuesExtractionQRIterative (double [] [] a, double accuracy, int maxIterations) takes the input along the matrix itself, as well as several more parameters:
At the output we get two matrices:
So let's start writing an algorithm, first we will create matrices that will contain eigenvectors and eigenvalues of the original matrix:
And run the loop until the algorithm stops:
Generate output:
And of course the test:
It should be noted that the eigenvectors can change direction from iteration to iteration, which will be seen by the change of the signs of their coordinates, but it is obvious that this is not significant.
Now everything is ready for the implementation of the article sabzh.
The hidden parameter of the model (class) will be a certain subset of the eigenvectors of the covariance matrix of the original data:
The constructor takes as input a dataset and dimension to which the feature space should be reduced, as well as parameters for QR decomposition. Inside, the covariance matrix is calculated and the first few eigenvectors are taken. In general, there is a way to choose the optimal number of eigenvectors for the desired information loss parameter, i.e. to which dimension you can reduce the space, while losing no more than a fixed percentage of information, but I omit this step, you can listen to it in the lecture of Andrew Ng on the course website.
Then we implement the direct and inverse transformation using the formulas from the beginning of the article.
To test, we will invent a small array of data, and checking the values in the lab (using the code from PCA = home), we will write a class for testing:
For my project, I needed the implementation of this method in c #, which I did today. The method of the main components is very elegant and beautiful, and if you do not understand the mathematics that lies behind all this, then it can all be called shamanism. The problem of calculating the eigenvectors of the matrix is that there is no quick way to calculate their exact values, so you have to get out. I want to talk about one of these ways to get out, as well as the c # code that performs this procedure. I ask under the cat.
Principal Component Method (No Code For Now)
')
Part one: algorithm initialization. The input to the algorithm is an array of data, as well as the dimension of the space to which it is necessary to reduce the data.
- The covariance matrix is calculated (this matrix has a remarkable property - it is symmetric, it will be very useful to us)
- Calculate the eigenvectors of the matrix
- The first en of eigenvectors are chosen, where en is the dimension to which the dimension of the feature space is to be reduced; selected vectors can be written vertically and assembled into a matrix
Part Two: Reducing the dimension of the input vector. At the input is a vector, dimensions as in the data array used in the initialization step; at the output, a vector of smaller dimension (or the projection of the input vector on an orthonormal basis formed by sampling from eigenvectors).
- By multiplying the scalar input vector by all the vectors from the sample of eigenvectors, we obtain a smaller vector:
Part three: recovery of the dimension of the vector (of course, with the loss of information). The input is a vector of dimension equal to that to which we reduced the vectors; output vector of the original dimension.
- If you transpose the matrix
 then there will be as many lines in it as the dimension of the input vector at the previous step, and the desired vector is restored by the formula:
then there will be as many lines in it as the dimension of the input vector at the previous step, and the desired vector is restored by the formula: 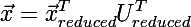
As you can see, the entire computational complexity is in the initialization step when calculating the covariance matrix and eigenvectors. I will not stop on the calculation of the covariance matrix, since it is calculated naively by definition. Having studied several algorithms for calculating the eigenvectors of the matrix, I stopped at the iterative QR decomposition method. He has a significant limitation, he gives a less accurate result only for symmetric matrices, but we were lucky :), the covariance matrix is just that. The second limitation of this algorithm is that the matrix must be full-rank .
QR decomposition
The iterative QR method for finding eigenvectors obviously uses QR decomposition , so first we have to implement this process. Gram-Schmidt algorithm was chosen to implement this process.
For this we need a function that calculates the projection of the vector a on the vector b :
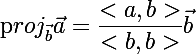 where <> - denotes the scalar product of vectors.
where <> - denotes the scalar product of vectors.public static double[] VectorProjection(double[] a, double[] b) { double k = ScalarVectorProduct(a, b)/ScalarVectorProduct(b, b); return ScalarToVectorProduct(k, b); } I will not dwell on even simpler functions, such as ScalarVectorProduct, so that the code size in the text does not exceed the critical limit.
So the actual QR decomposition procedure itself IList <double []> DecompositionGS (double [] [] a) receives a matrix as input, and returns two matrices in response:
- The first contains in its columns an orthonormal basis such that
 ;
; - the second matrix will be upper triangular .
First, we divide the matrix into columns and write them into the av list:
List<double[]> av = new List<double[]>(); foreach (double[] vector in DecomposeMatrixToColumnVectors(a)) { av.Add(vector); } We initialize two auxiliary lists:
List<double[]> u = new List<double[]>(); u.Add(av[0]); List<double[]> e = new List<double[]>(); e.Add(ScalarToVectorProduct(1 / NormOfVector(u[0]), u[0])); These are the same lists used in the algorithm scheme:

After initialization, the first u and e have been calculated, we continue in the loop to calculate the following values:
for (int i = 1; i < a.Length; i++) { double[] projAcc = new double[a.Length]; for (int j = 0; j < projAcc.Length; j++) { projAcc[j] = 0; } for (int j = 0; j < i; j++) { double[] proj = VectorProjection(av[i], e[j]); for (int k = 0; k < projAcc.Length; k++) { projAcc[k] += proj[k]; } } double[] ui = new double[a.Length]; for (int j = 0; j < ui.Length; j++) { ui[j] = a[j][i] - projAcc[j]; } u.Add(ui); e.Add(ScalarToVectorProduct(1/NormOfVector(u[i]), u[i])); } Finally, we generate data in the output format:
double[][] q = new double[a.Length][]; for (int i = 0; i < q.Length; i++) { q[i] = new double[a.Length]; for (int j = 0; j < q[i].Length; j++) { q[i][j] = e[j][i]; } } double[][] r = new double[a.Length][]; for (int i = 0; i < r.Length; i++) { r[i] = new double[a.Length]; for (int j = 0; j < r[i].Length; j++) { if (i >= j) { r[i][j] = ScalarVectorProduct(e[j], av[i]); } else { r[i][j] = 0; } } } r = Transpose(r); List<double[][]> res = new List<double[][]>(); res.Add(q); res.Add(r); return res; Hooray! Now you need a test for this:
[Test(Description = "Test of QRDecompositionGS")] public void QRDecompositionGSTest() { double[][] a = new double[][] { new double[] {12, -51, 4}, new double[] {6, 167, -68}, new double[] {-4, 24, -41} }; IList<double[][]> res = LinearAlgebra.QRDecompositionGS(a); double[][] expQ = new double[][] { new double[] {6.0/7, -69.0/175, -58.0/175}, new double[] {3.0/7, 158.0/175, 6.0/175}, new double[] {-2.0/7, 6.0/35, -33.0/35} }; double[][] expR = new double[][] { new double[] {14, 21, -14}, new double[] {0, 175, -70}, new double[] {0, 0, 35} }; Assert.True(Helper.AreEqualMatrices(expQ, res[0], 0.0001), "expQ != Q"); Assert.True(Helper.AreEqualMatrices(expR, res[1], 0.0001), "expR != R"); } The Helper.AreEqualMatrices function compares element-wise matrices with the accuracy of the third parameter.
Iterative QR Method
The iterative QR method is based on the remarkable theorem, the essence of which is that if you initialize the matrix A zero as
and then multiply all obtained Q , then the result will be a matrix, in the columns of which will be the eigenvectors of the original matrix, the values of which will be the more accurate, the longer the process was performed. In other words, when the number of iterations tends to infinity, the product
So, the QR iteration method IList <double [] []> EigenVectorValuesExtractionQRIterative (double [] [] a, double accuracy, int maxIterations) takes the input along the matrix itself, as well as several more parameters:
- double accuracy - the accuracy of which we want to achieve, the algorithm will stop if the changes in the values of the eigenvectors are no more than this value;
- int maxIterations - the maximum number of iterations.
At the output we get two matrices:
- the first contains in its columns the eigenvectors of the matrix a ;
- The second matrix on its main diagonal contains the eigenvalues of the matrix a .
So let's start writing an algorithm, first we will create matrices that will contain eigenvectors and eigenvalues of the original matrix:
double[][] aItr = a; double[][] q = null; And run the loop until the algorithm stops:
for (int i = 0; i < maxIterations; i++) { IList<double[][]> qr = QRDecompositionGS(aItr); aItr = MatricesProduct(qr[1], qr[0]); if (q == null) { q = qr[0]; } else { double[][] qNew = MatricesProduct(q, qr[0]); bool accuracyAcheived = true; for (int n = 0; n < q.Length; n++) { for (int m = 0; m < q[n].Length; m++) { if (Math.Abs(Math.Abs(qNew[n][m]) - Math.Abs(q[n][m])) > accuracy) { accuracyAcheived = false; break; } } if (!accuracyAcheived) { break; } } q = qNew; if (accuracyAcheived) { break; } } } Generate output:
List<double[][]> res = new List<double[][]>(); res.Add(q); res.Add(aItr); return res; And of course the test:
[Test(Description = "Test of Eigen vectors extraction")] public void EigenVectorExtraction() { double[][] a = new double[][] { new double[] {1, 2, 4}, new double[] {2, 9, 8}, new double[] {4, 8, 2} }; IList<double[][]> ev = LinearAlgebra.EigenVectorValuesExtractionQRIterative(a, 0.001, 1000); double expEV00 = 15.2964; double expEV11 = 4.3487; double expEV22 = 1.0523; Assert.AreEqual(expEV00, Math.Round(Math.Abs(ev[1][0][0]), 4)); Assert.AreEqual(expEV11, Math.Round(Math.Abs(ev[1][1][1]), 4)); Assert.AreEqual(expEV22, Math.Round(Math.Abs(ev[1][2][2]), 4)); } It should be noted that the eigenvectors can change direction from iteration to iteration, which will be seen by the change of the signs of their coordinates, but it is obvious that this is not significant.
Implementation of the principal component method
Now everything is ready for the implementation of the article sabzh.
The hidden parameter of the model (class) will be a certain subset of the eigenvectors of the covariance matrix of the original data:
private IList<double[]> _eigenVectors = null; The constructor takes as input a dataset and dimension to which the feature space should be reduced, as well as parameters for QR decomposition. Inside, the covariance matrix is calculated and the first few eigenvectors are taken. In general, there is a way to choose the optimal number of eigenvectors for the desired information loss parameter, i.e. to which dimension you can reduce the space, while losing no more than a fixed percentage of information, but I omit this step, you can listen to it in the lecture of Andrew Ng on the course website.
internal DimensionalityReductionPCA(IList<double[]> dataSet, double accuracyQR, int maxIterationQR, int componentsNumber) { double[][] cov = BasicStatFunctions.CovarianceMatrixOfData(dataSet); IList<double[][]> eigen = LinearAlgebra.EigenVectorValuesExtractionQRIterative(cov, accuracyQR, maxIterationQR); IList<double[]> eigenVectors = LinearAlgebra.DecomposeMatrixToColumnVectors(eigen[0]); if (componentsNumber > eigenVectors.Count) { throw new ArgumentException("componentsNumber > eigenVectors.Count"); } _eigenVectors = new List<double[]>(); for (int i = 0; i < componentsNumber; i++) { _eigenVectors.Add(eigenVectors[i]); } } Then we implement the direct and inverse transformation using the formulas from the beginning of the article.
public double[] Transform(double[] dataItem) { if (_eigenVectors[0].Length != dataItem.Length) { throw new ArgumentException("_eigenVectors[0].Length != dataItem.Length"); } double[] res = new double[_eigenVectors.Count]; for (int i = 0; i < _eigenVectors.Count; i++) { res[i] = 0; for (int j = 0; j < dataItem.Length; j++) { res[i] += _eigenVectors[i][j]*dataItem[j]; } } return res; } public double[] Reconstruct(double[] transformedDataItem) { if (_eigenVectors.Count != transformedDataItem.Length) { throw new ArgumentException("_eigenVectors.Count != transformedDataItem.Length"); } double[] res = new double[_eigenVectors[0].Length]; for (int i = 0; i < res.Length; i++) { res[i] = 0; for (int j = 0; j < _eigenVectors.Count; j++) { res[i] += _eigenVectors[j][i]*transformedDataItem[j]; } } return res; } Testing
To test, we will invent a small array of data, and checking the values in the lab (using the code from PCA = home), we will write a class for testing:
[TestFixture(Description = "Test of DimensionalityReductionPCA")] public class DimensionalityReductionPCATest { private IList<double[]> _data = null; private IDataTransformation<double[]> _transformation = null; private double[] _v = null; [SetUp] public void SetUp() { _v = new double[] { 1, 0, 3 }; _data = new List<double[]>() { new double[] {1, 2, 23}, new double[] {-3, 17, 5}, new double[] {13, -6, 7}, new double[] {7, 8, -9} }; _transformation = new DimensionalityReductionPCA(_data, 0.0001, 1000, 2); } [Test(Description = "Test of DimensionalityReductionPCA transform")] public void DimensionalityReductionPCATransformTest() { double[] reduced = _transformation.Transform(_v); double[] expReduced = new double[] {-2.75008, 0.19959}; Assert.IsTrue(Helper.AreEqualVectors(expReduced, reduced, 0.001)); double[] reconstructed = _transformation.Reconstruct(reduced); double[] expReconstructed = new double[] {-0.21218, -0.87852, 2.60499}; Assert.IsTrue(Helper.AreEqualVectors(expReconstructed, reconstructed, 0.001)); } Links
- course on machine learning
- an article that describes an iterative QR method
- a bunch of links to wikipedia in the text of the article
Source: https://habr.com/ru/post/146236/
All Articles