Bioinformatics: a view from the inside
 Of all the technical and natural sciences known to me, it is perhaps the worst idea about bioinformatics that people have. It is either incorrect to some degree or not at all. When two years ago I started studying bionformatics, I myself didn’t have any knowledge about this science. Over time, I began to better understand what tasks bioinformatics are facing, what they use, and what may be the result of their work.
Of all the technical and natural sciences known to me, it is perhaps the worst idea about bioinformatics that people have. It is either incorrect to some degree or not at all. When two years ago I started studying bionformatics, I myself didn’t have any knowledge about this science. Over time, I began to better understand what tasks bioinformatics are facing, what they use, and what may be the result of their work.Bioinformatics have no test tubes, reagents, bacteria, white coats. The main tools they have are a laptop, a pen with paper or a white board with a marker - in general, everything is like that of a programmer. And the work itself is very similar to the work in an IT company, and the laboratory is like a small development department. What are the differences then? Well, I'll try to answer.
Firstly, the tasks are mainly algorithmic. That is, before you write a program, you need to read a few articles, think for yourself, discuss your ideas with colleagues and only then proceed to implement. Secondly, we have to work with large amounts of data, and therefore the implementation should be as efficient as possible. However, even an effective, logical and perfectly debugged program may not give the desired result. The main reason for this is the biological origin of the data, which means a huge number of errors and a significant difference between the data from different laboratories.
')
Another, perhaps the most visible difference between bioinformatics and programming is research and publication. Bioinformatics is a science, which means you just need to be aware of everything that happens in the world. To this end, there are numerous conferences, collaborations with laboratories from other countries, and, of course, publications — you also need to tell everyone about your achievements. Without all this, you can diligently reinvent the wheel.
In general, the impression of bioinformatics is exactly that, but it is best to tell it with an example, especially since there is one, and very close. But first things first.
Laboratory of Algorithmic Biology
In 2010, the program “megagrants” was launched in Russia. Under the leadership of leading Western scientists (in most cases, who had long since left Russia), new scientific laboratories were created. The laboratory of algorithmic biology at SPbAU under the leadership of Pavel Pevzner, one of the most famous scientists in his field, became one of such. Pavel graduated from Moscow Institute of Physics and Technology, but soon after that he left for the USA, took up Computer Science (or, to be precise, it was bioinformatics) and is now a professor at the University of California at San Diego.
Before you tell what exactly they are doing in the laboratory, it is worthwhile to introduce readers to the current situation.
Little about genomics
I am sure that every reader has heard the word genome. For biologists, the genome is DNA molecules - long chains consisting of four nucleotides, organized into chromosomes, folded in the nucleus of a cell. We see the genome as a string, again consisting of four characters (A, C, G, T). The length of the genome can reach billions or even tens of billions of characters. Biologists do not know how to read the entire genome - only small fragments up to 150 "letters", and even then with errors. Our task is to restore the original genome for these pieces, or as they often say, to collect.
For clarity, you can make such a comparison: imagine a pack of identical newspapers. Now imagine that this pack is blown up and small pieces of paper fly away, shuffle, deteriorate or even burn completely. And then on the heap of this garbage I want to glue the original newspaper.
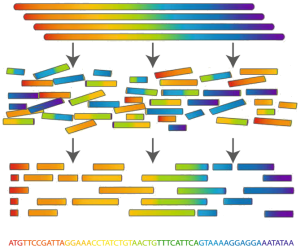 The same with the genome. The first technologies made it possible to read pieces of the genome with a length of up to several thousand characters. These technologies were incredibly expensive — several billion dollars were spent on assembling the first human genome and several years of hard work by hundreds of laboratory workers around the world. Modern technologies allow you to read shorter fragments, but much cheaper and in large quantities. Processing gigabytes of input data, of course, automatically. For this purpose, programs are developed that are called genomic collectors, or more often assemblers (assembled from English). Due to some features of the original genomes (for example, recurring regions), as well as a large number of errors in the input data, the result of the work of the collector is not a whole gene, but rather long enough parts of it. The longer the resulting sites, the more they are similar to the original genome, the better the result is considered.
The same with the genome. The first technologies made it possible to read pieces of the genome with a length of up to several thousand characters. These technologies were incredibly expensive — several billion dollars were spent on assembling the first human genome and several years of hard work by hundreds of laboratory workers around the world. Modern technologies allow you to read shorter fragments, but much cheaper and in large quantities. Processing gigabytes of input data, of course, automatically. For this purpose, programs are developed that are called genomic collectors, or more often assemblers (assembled from English). Due to some features of the original genomes (for example, recurring regions), as well as a large number of errors in the input data, the result of the work of the collector is not a whole gene, but rather long enough parts of it. The longer the resulting sites, the more they are similar to the original genome, the better the result is considered.Genome assembly task
If we take the task of assembling a genome in the most general case, it will be nothing more than the shortest superstring problem, which is formulated as follows: find the shortest string such that each line in the given set would be its substring. This task is NP-complete. But if we assume that we have all the possible substrings of the source string of the same length, the problem can be solved in polynomial time. Assembly of the genome is just such a case. In 2001, Pavel Pevzner proposed an efficient genome assembly approach using the Count de Bruin. The main idea of this approach is used in almost every modern genomic assembler. However, in practice, everything is greatly complicated by the aforementioned biological errors, and therefore the main task is to develop heuristics for various subtasks arising during the assembly of genomes.
In the laboratory of algorithmic biology, it was decided to focus on the development of the assembler. Of course, at the time of the creation of the laboratory there was a huge number of genomic collectors. Why was it to create another one? In fact, the assembly task turns out to be much wider than it seems at first glance. Biologists produce a huge number of different types of input data, each of which requires the development of new methods that take into account their specifics. In addition, the assembly of the genome includes a large number of stages and algorithms, so even despite the fact that all modern assemblers use the same approach, their results can be very different. The laboratory was tasked with obtaining an assembler that would surpass existing ones in many ways.
Path to bioinformatics
I got into bioinformatics, one might say, by chance. I studied at the SPbAU Master's program and, like every student, at the beginning of the semester I had to choose a research paper. To try myself in a new field, I chose a bioinformatics project. At first, the scarecrow is that you may have to learn biology, instead of developing and implementing algorithms. However, the fears, fortunately, were not justified - immersion in this subject area is exactly the same as in any other. Gradually, you begin to understand more, learn something new, and even if biology was far from being the most favorite subject in school, interest in it appears fairly quickly. Almost immediately, I realized that bioinformatics is exactly what I wanted to do - programming with elements of research and an interesting subject area.
While I was working on my project, the laboratory of algorithmic biology I mentioned was organized. In the summer of 2011, I successfully completed my internship in her and remained as a permanent research assistant. If we talk about the laboratory as a whole - a huge number of various interesting projects that are not limited to assembling genomes, cooperation with Western laboratories, scientific conferences, a constant opportunity to learn something new and, of course, a very good team.
One could probably talk for a long time about both work in the laboratory and bioinformatics in general, which still has a lot of open problems, and concrete approaches and algorithms for various tasks. But it is impossible to embrace the immensity, and therefore the story will be about one thing and the next time. And what exactly it depends on your wishes.
Links
Source: https://habr.com/ru/post/143115/
All Articles