Part number 6. Introduction to Folding RNA
So, in the last parts we figured out how to comparatively simply fold the helix of RNA. Now we have to understand how RNA generally folds. The RNA that we took as an example has three helices. Two of them L1 and L2 can be minimized independently. But with the third problem. This third consists of RNA ends, and when it is folded, our folded helixes L1 and L2 begin to move. First, they interfere with each other, and therefore the folding of the third helix. Secondly, the formation of about a dozen different pseudo-symmetric structures is possible - the L1, L2 helixs can be arranged differently with respect to the folding ends of RNA.
Here we will try to figure out how to solve these problems.
')
Repeat drawing
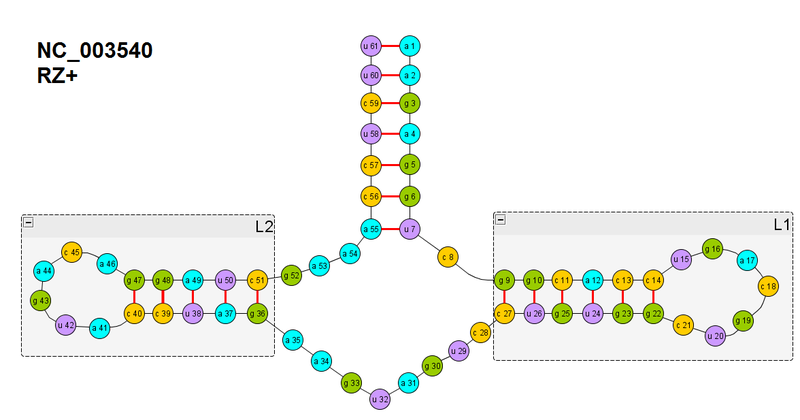
As I wrote in the last part in the comments, this is a Chrysanthemum chlorotic mottle viroid viroid ribozyme (NC_003540). Only its primary sequence has been obtained, but there is no tertiary one yet. But we will try to get it in the computer model in silico.
The secondary structure (hydrogen bonds between nucleotides) is well predicted by a number of already existing programs, for example, here . But what is important! They predict hydrogen bonds only spirals. But the fact is that in order for the spirals to be held together there are inter- spiral hydrogen bonds.
How to recognize them? But we know that ribozymes have a close (conservative) structure in some areas. Take the one that already exists in the PDB databank. For example, satellite tobacco ringspot virus hammerhead RNA ( 2QUS )
It was obtained by X-ray crystallography, and there, alas, there are no hydrogen atoms, and no hydrogen bonds are visible. But if you look in more detail (to measure the distances between certain atoms), then you can quite easily find out where these hydrogen bonds should be. We are not interested in the standard ones, so we are looking for those who tighten the spirals together and other non-standard ones. And we find.
They are between nucleotides a46-u19, a46-u24, c32-g37.
Well, but to what pairs of nucleotides - does this correspond in the ribozyme of interest to us (NC_003540)?
Here it is necessary to make a so-called. alignment There are two primary sequences:
1. (NC_003540) aagaggucggcaccugacgucgguguccugaugaagauccaugacaggaucgaaaccucuu
2. (2QUS in PDB) gggagcccugucaccggaugugcuuuccggucugaugaguccgugggacaaaacagggcucccgaauu
And aligning them is not so trivial. Here I will ask the experts who read me to do it. And then we compare the results. I argue that modern programs that do the comparison automatically do not cope with it. Let's check?
Further suppose we have found what intercoiled hydrogen bonds need to form. And here we are waiting for the next problem. And the problem is that we believed the hypothesis, according to which the secondary structures are first collapsed and only then the subsequent collapsing occurs (the so-called Hierarchical model). This model is incorrect, at least in order to use it in computer calculations.
Why? The fact is that the formation of a spiral rigidly connects the position of the loop (the freedom of nucleotides in the loop becomes very small). There are many different kinds of loops. In our case, where the criterion is only the formation of hydrogen bonds of the helix - the loop is almost random. And we need to loop the spiral L1 most closely packed with a loop L2. If this does not happen, we will not form the inter-helical bonds required by us. Just the necessary atoms will not be able to come close enough for the formation of hydrogen bonds - already curtailed spirals will interfere.
But this is not all the difficulties. In the last part in the comments, I showed how the hydrogen bonds between the spirals may come from the wrong side, but from the other side, and everything will be fine. Only the spiral L3 will not be able to form later.
But what is needed for the spirals L1 and L2 to come together on the right side? It turns out that not everything can solve hydrogen bonds. An important influence has a so-called. interaction stacking . But if we take into account these interactions between all nucleotides, then we will spend too much time.
In addition, I did not find a sufficiently accurate mathematical description of the interaction stacking (similar to how I described hydrogen bonds). Can someone tell me?
In any case, we find the interaction that is crucial for folding the stacking, and then our spirals approach each other from the right side.
Well, that's all the technology. Then just practice.
Previous parts From proteins to RNA , Mat. criteria How to reduce the number of turns the chain? How to evaluate the course of folding of single-stranded RNA? , The limitation of optimizing methods in games with the opponent and without , One fundamental problem
Here we will try to figure out how to solve these problems.
')
Repeat drawing

As I wrote in the last part in the comments, this is a Chrysanthemum chlorotic mottle viroid viroid ribozyme (NC_003540). Only its primary sequence has been obtained, but there is no tertiary one yet. But we will try to get it in the computer model in silico.
The secondary structure (hydrogen bonds between nucleotides) is well predicted by a number of already existing programs, for example, here . But what is important! They predict hydrogen bonds only spirals. But the fact is that in order for the spirals to be held together there are inter- spiral hydrogen bonds.
How to recognize them? But we know that ribozymes have a close (conservative) structure in some areas. Take the one that already exists in the PDB databank. For example, satellite tobacco ringspot virus hammerhead RNA ( 2QUS )
It was obtained by X-ray crystallography, and there, alas, there are no hydrogen atoms, and no hydrogen bonds are visible. But if you look in more detail (to measure the distances between certain atoms), then you can quite easily find out where these hydrogen bonds should be. We are not interested in the standard ones, so we are looking for those who tighten the spirals together and other non-standard ones. And we find.
They are between nucleotides a46-u19, a46-u24, c32-g37.
Well, but to what pairs of nucleotides - does this correspond in the ribozyme of interest to us (NC_003540)?
Here it is necessary to make a so-called. alignment There are two primary sequences:
1. (NC_003540) aagaggucggcaccugacgucgguguccugaugaagauccaugacaggaucgaaaccucuu
2. (2QUS in PDB) gggagcccugucaccggaugugcuuuccggucugaugaguccgugggacaaaacagggcucccgaauu
And aligning them is not so trivial. Here I will ask the experts who read me to do it. And then we compare the results. I argue that modern programs that do the comparison automatically do not cope with it. Let's check?
Further suppose we have found what intercoiled hydrogen bonds need to form. And here we are waiting for the next problem. And the problem is that we believed the hypothesis, according to which the secondary structures are first collapsed and only then the subsequent collapsing occurs (the so-called Hierarchical model). This model is incorrect, at least in order to use it in computer calculations.
Why? The fact is that the formation of a spiral rigidly connects the position of the loop (the freedom of nucleotides in the loop becomes very small). There are many different kinds of loops. In our case, where the criterion is only the formation of hydrogen bonds of the helix - the loop is almost random. And we need to loop the spiral L1 most closely packed with a loop L2. If this does not happen, we will not form the inter-helical bonds required by us. Just the necessary atoms will not be able to come close enough for the formation of hydrogen bonds - already curtailed spirals will interfere.
But this is not all the difficulties. In the last part in the comments, I showed how the hydrogen bonds between the spirals may come from the wrong side, but from the other side, and everything will be fine. Only the spiral L3 will not be able to form later.
But what is needed for the spirals L1 and L2 to come together on the right side? It turns out that not everything can solve hydrogen bonds. An important influence has a so-called. interaction stacking . But if we take into account these interactions between all nucleotides, then we will spend too much time.
In addition, I did not find a sufficiently accurate mathematical description of the interaction stacking (similar to how I described hydrogen bonds). Can someone tell me?
In any case, we find the interaction that is crucial for folding the stacking, and then our spirals approach each other from the right side.
Well, that's all the technology. Then just practice.
Previous parts From proteins to RNA , Mat. criteria How to reduce the number of turns the chain? How to evaluate the course of folding of single-stranded RNA? , The limitation of optimizing methods in games with the opponent and without , One fundamental problem
Source: https://habr.com/ru/post/140991/
All Articles