Overview of data clustering algorithms
Greetings
In my thesis, I conducted a review and comparative analysis of data clustering algorithms. I thought that already collected and developed material may be interesting and useful to someone.
Sashaeve described the article “Clustering: k-means and c-means algorithms” about clustering . I will partly repeat the words of Alexander, partially add. Also at the end of this article, those interested can read the materials on the links in the reference list.
I also tried to bring a dry "diploma" style of presentation to a more journalistic.
Clustering (or cluster analysis) is the task of breaking up a set of objects into groups called clusters. Inside each group there should be “similar” objects, and the objects of different groups should be as different as possible. The main difference between clustering and classification is that the list of groups is not clearly defined and is determined during the operation of the algorithm.
')
The use of cluster analysis in general is reduced to the following steps:
After receiving and analyzing the results, it is possible to adjust the selected metric and the clustering method to an optimal result.
So, how to determine the "similarity" of objects? First you need to make a vector of characteristics for each object - as a rule, it is a set of numerical values, for example, height and weight of a person. However, there are also algorithms that work with qualitative (so-called categorical) characteristics.
After we have determined the vector of characteristics, it is possible to carry out a normalization so that all components give the same contribution when calculating the “distance”. In the normalization process, all values are reduced to a certain range, for example, [-1, -1] or [0, 1].
Finally, for each pair of objects, the “distance” between them is measured - the degree of similarity. There are many metrics, here are just the main ones:
The choice of the metric lies entirely with the researcher, since the results of clusterization can differ significantly when using different measures.
For myself, I singled out two main classifications of clustering algorithms.
In the case of using hierarchical algorithms, the question arises how to combine the clusters among themselves, how to calculate the “distances” between them. There are several metrics:
Among the algorithms of hierarchical clustering, there are two main types: ascending and descending algorithms. Top-down algorithms work on the “top-down” principle: at the beginning, all objects are placed in one cluster, which is then divided into smaller and smaller clusters. Ascending algorithms are more common, which at the beginning of the work place each object in a separate cluster, and then merge clusters into ever larger ones until all the objects in the sample are contained in one cluster. Thus, a system of nested partitions is built. The results of such algorithms are usually presented in the form of a tree - dendrograms. A classic example of such a tree is the classification of animals and plants.
To calculate the distances between clusters, more often everyone uses two distances: a single bond or a full bond (see an overview of the measures of the distances between clusters).
The lack of hierarchical algorithms can include a system of complete partitions, which may be superfluous in the context of the problem being solved.
The task of clustering can be viewed as the construction of an optimal partitioning of objects into groups. At the same time, optimality can be defined as the requirement to minimize the root-mean-square split error:
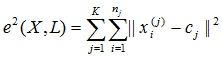
where c j is the “center of mass” of the cluster j (a point with average values of characteristics for a given cluster).
The quadratic error algorithms are of the type of flat algorithms. The most common algorithm in this category is the k-means method. This algorithm builds a given number of clusters located as far as possible from each other. The operation of the algorithm is divided into several stages:
As a criterion for stopping the operation of the algorithm, usually choose the minimum change in the root mean square error. It is also possible to stop the operation of the algorithm, if at step 2 there were no objects that moved from cluster to cluster.
The disadvantages of this algorithm include the need to specify the number of clusters for splitting.
The most popular fuzzy clustering algorithm is the c-means algorithm. It is a modification of the k-means method. Algorithm steps:
This algorithm may not be suitable if the number of clusters is not known in advance, or it is necessary to clearly assign each object to one cluster.
The essence of such algorithms is that the sample of objects is represented as a graph G = (V, E) , whose vertices correspond to objects, and the edges have a weight equal to the “distance” between the objects. The advantages of graph clustering algorithms are visibility, relative ease of implementation, and the ability to make various improvements based on geometrical considerations. The main algorithms are the algorithm for selecting connected components, the algorithm for constructing the minimum spanning (spanning) tree and the layer-by-layer clustering algorithm.
In the algorithm for selecting connected components, the input parameter R is specified and all edges for which “distances” are greater than R are removed in the graph. Only the closest pairs of objects remain connected. The meaning of the algorithm is to find a value of R that lies in the range of all “distances” at which the graph “falls apart” into several connected components. The resulting components are clusters.
For the selection of the parameter R , a histogram of distributions of pairwise distances is usually constructed. In problems with a well-pronounced cluster data structure, there will be two peaks in the histogram — one corresponds to intracluster distances, and the second to intercluster distances. The parameter R is selected from the minimum zone between these peaks. At the same time, managing the number of clusters using the distance threshold is rather difficult.
The minimum spanning tree algorithm first builds a minimal spanning tree on the graph, and then successively removes the edges with the greatest weight. The figure shows the minimum spanning tree obtained for nine objects.
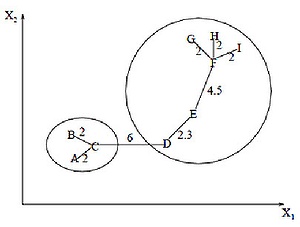
By removing the link labeled CD, with a length of 6 units (edge with maximum distance), we obtain two clusters: {A, B, C} and {D, E, F, G, H, I}. The second cluster can be further divided into two more clusters by removing the edge EF, which has a length of 4.5 units.
The layer-by-layer clustering algorithm is based on identifying connected components of a graph at some level of distance between objects (vertices). The distance level is set by the distance threshold c . For example, if the distance between objects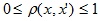 then
then 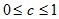 .
.
The layer clustering algorithm generates a sequence of subgraphs of the graph G , which reflect the hierarchical relationships between clusters:
 ,
,
where G t = (V, E t ) is a graph at the level of t ,
 ,
,
with t - t-th threshold of distance,
m is the number of hierarchy levels,
G 0 = (V, o) , o is the empty set of edges of the graph obtained when t 0 = 1,
G m = G , that is, the graph of objects without restrictions on the distance (the length of the edges of the graph), since t m = 1.
By changing the distance thresholds { c 0 , ..., m }, where 0 = c 0 < c 1 <... < c m = 1, it is possible to control the depth of the hierarchy of the resulting clusters. Thus, a layer-by-layer clustering algorithm is able to create both flat data partitioning and hierarchical.
Computational complexity of algorithms
Comparative table of algorithms
In my work, I needed to select separate areas from the hierarchical structures (trees). Those. in fact, it was necessary to cut the original tree into several smaller trees. Since an oriented tree is a special case of a graph, algorithms based on graph theory are naturally appropriate.
In contrast to a fully connected graph, in an oriented tree, not all vertices are connected by edges, and the total number of edges is n – 1, where n is the number of vertices. Those. applied to the nodes of the tree, the operation of the algorithm for selecting connected components will be simplified, since the removal of any number of edges will “decompose” the tree into connected components (individual trees). The algorithm of the minimum spanning tree in this case will coincide with the algorithm for selecting connected components — by removing the longest edges, the original tree is divided into several trees. It is obvious that the construction phase of the minimal spanning tree itself is skipped.
In the case of using other algorithms, they would have to separately consider the presence of connections between objects, which complicates the algorithm.
Separately, I want to say that in order to achieve the best result, it is necessary to experiment with the choice of distance measures, and sometimes even change the algorithm. There is no single solution.
1. Vorontsov K.V. Algorithms for clustering and multidimensional scaling . Lecture course. Moscow State University, 2007.
2. Jain A., Murty M., Flynn P. Data Clustering: A Review . // ACM Computing Surveys. 1999. Vol. 31, no. 3
3. Kotov A., Krasilnikov N. Data clustering . 2006
3. Mandel I. D. Cluster analysis. - M .: Finance and Statistics, 1988.
4. Applied statistics: classification and reduction of dimension. / S.A. Ayvazyan, V.M. Buchstaber, I.S. Enyukov, LD Meshalkin - M .: Finance and Statistics, 1989.
5. Information and analytical resource dedicated to machine learning, pattern recognition and data mining - www.machinelearning.ru
6. Chubukova I.A. Course of lectures "Data Mining", Internet University of Information Technologies - www.intuit.ru/department/database/datamining
In my thesis, I conducted a review and comparative analysis of data clustering algorithms. I thought that already collected and developed material may be interesting and useful to someone.
Sashaeve described the article “Clustering: k-means and c-means algorithms” about clustering . I will partly repeat the words of Alexander, partially add. Also at the end of this article, those interested can read the materials on the links in the reference list.
I also tried to bring a dry "diploma" style of presentation to a more journalistic.
Clustering concept
Clustering (or cluster analysis) is the task of breaking up a set of objects into groups called clusters. Inside each group there should be “similar” objects, and the objects of different groups should be as different as possible. The main difference between clustering and classification is that the list of groups is not clearly defined and is determined during the operation of the algorithm.
')
The use of cluster analysis in general is reduced to the following steps:
- Selecting a selection of objects for clustering.
- Determining the set of variables by which the objects in the sample will be evaluated. If necessary, normalize the values of variables.
- Calculating the values of the similarity measure between objects.
- The use of cluster analysis to create groups of similar objects (clusters).
- Presentation of analysis results.
After receiving and analyzing the results, it is possible to adjust the selected metric and the clustering method to an optimal result.
Distance measures
So, how to determine the "similarity" of objects? First you need to make a vector of characteristics for each object - as a rule, it is a set of numerical values, for example, height and weight of a person. However, there are also algorithms that work with qualitative (so-called categorical) characteristics.
After we have determined the vector of characteristics, it is possible to carry out a normalization so that all components give the same contribution when calculating the “distance”. In the normalization process, all values are reduced to a certain range, for example, [-1, -1] or [0, 1].
Finally, for each pair of objects, the “distance” between them is measured - the degree of similarity. There are many metrics, here are just the main ones:
- Euclidean distance
The most common distance function. It is a geometric distance in multidimensional space: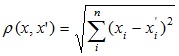
- Square euclidean distance
It is used to give more weight to more distant objects. This distance is calculated as follows:
- City block distance (Manhattan distance)
This distance is the average difference in coordinates. In most cases, this measure of distance leads to the same results as for the usual Euclidean distance. However, for this measure, the influence of individual large differences (emissions) is reduced (since they are not squared). The formula for calculating the Manhattan distance is:
- Chebyshev distance
This distance can be useful when you need to define two objects as “different” if they differ in any one coordinate. Chebyshev distance is calculated by the formula:
- Power distance
It is used when it is necessary to increase or decrease the weight related to the dimension for which the corresponding objects are very different. The power distance is calculated using the following formula: ,
,
where r and p are user-defined parameters. The parameter p is responsible for the gradual weighting of differences in individual coordinates, the parameter r is responsible for the progressive weighing of large distances between objects. If both parameters - r and p - are equal to two, then this distance coincides with the Euclidean distance.
The choice of the metric lies entirely with the researcher, since the results of clusterization can differ significantly when using different measures.
Algorithm classification
For myself, I singled out two main classifications of clustering algorithms.
- Hierarchical and flat.
Hierarchical algorithms (also called taxonomy algorithms) do not build a single partitioning of a sample into disjoint clusters, but a system of nested partitions. So at the output we get a cluster tree, the root of which is the entire sample, and the leaves are the smallest clusters.
Planar algorithms build one partitioning of objects into clusters. - Clear and fuzzy.
Clear (or non-intersecting) algorithms for each sample object are assigned a cluster number, i.e. each object belongs to only one cluster. Fuzzy (or intersecting) algorithms for each object associate a set of real values, indicating the degree of the object's relation to the clusters. Those. each object belongs to each cluster with a certain probability.
Clustering
In the case of using hierarchical algorithms, the question arises how to combine the clusters among themselves, how to calculate the “distances” between them. There are several metrics:
- Single connection (nearest neighbor distance)
In this method, the distance between two clusters is determined by the distance between the two closest objects (nearest neighbors) in different clusters. The resulting clusters tend to chain together. - Full communication (distance of the most distant neighbors)
In this method, the distances between clusters are determined by the largest distance between any two objects in different clusters (that is, the most distant neighbors). This method usually works very well when objects come from separate groups. If the clusters are elongated or their natural type is “chained,” then this method is unsuitable. - Unweighted pairwise mean
In this method, the distance between two different clusters is calculated as the average distance between all pairs of objects in them. The method is effective when objects form different groups, but it works equally well in the case of extended (“chained” type) clusters. - Weighted paired average
The method is identical to the unweighted pairwise average method, except that the size of the corresponding clusters (that is, the number of objects contained in them) is used as a weighting factor in the calculations. Therefore, this method should be used when unequal cluster sizes are assumed. - Unweighted centroid method
In this method, the distance between two clusters is defined as the distance between their centers of gravity. - Weighted centroid method (median)
This method is identical to the previous one, except that the calculations use weights to account for the difference between cluster sizes. Therefore, if there are or are suspected significant differences in cluster sizes, this method is preferable to the previous one.
Algorithm Overview
Hierarchical clustering algorithms
Among the algorithms of hierarchical clustering, there are two main types: ascending and descending algorithms. Top-down algorithms work on the “top-down” principle: at the beginning, all objects are placed in one cluster, which is then divided into smaller and smaller clusters. Ascending algorithms are more common, which at the beginning of the work place each object in a separate cluster, and then merge clusters into ever larger ones until all the objects in the sample are contained in one cluster. Thus, a system of nested partitions is built. The results of such algorithms are usually presented in the form of a tree - dendrograms. A classic example of such a tree is the classification of animals and plants.
To calculate the distances between clusters, more often everyone uses two distances: a single bond or a full bond (see an overview of the measures of the distances between clusters).
The lack of hierarchical algorithms can include a system of complete partitions, which may be superfluous in the context of the problem being solved.
Quadratic error algorithms
The task of clustering can be viewed as the construction of an optimal partitioning of objects into groups. At the same time, optimality can be defined as the requirement to minimize the root-mean-square split error:
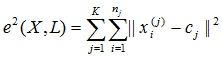
where c j is the “center of mass” of the cluster j (a point with average values of characteristics for a given cluster).
The quadratic error algorithms are of the type of flat algorithms. The most common algorithm in this category is the k-means method. This algorithm builds a given number of clusters located as far as possible from each other. The operation of the algorithm is divided into several stages:
- Randomly select k points, which are the initial "centers of mass" of the clusters.
- Assign each object to a cluster with the nearest "center of mass".
- Recalculate the "centers of mass" clusters according to their current composition.
- If the criterion for stopping the algorithm is not satisfied, return to step 2.
As a criterion for stopping the operation of the algorithm, usually choose the minimum change in the root mean square error. It is also possible to stop the operation of the algorithm, if at step 2 there were no objects that moved from cluster to cluster.
The disadvantages of this algorithm include the need to specify the number of clusters for splitting.
Fuzzy algorithms
The most popular fuzzy clustering algorithm is the c-means algorithm. It is a modification of the k-means method. Algorithm steps:
- Select the initial fuzzy partition of n objects into k clusters by choosing the membership matrix U of size nxk .
- Using the matrix U, find the value of the fuzzy error criterion:
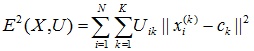 ,
,
where c k is the “center of mass” of a fuzzy cluster k :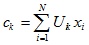 .
. - Regroup objects in order to reduce this value of the fuzzy error criterion.
- Return to Section 2 until the changes in the matrix U become insignificant.
This algorithm may not be suitable if the number of clusters is not known in advance, or it is necessary to clearly assign each object to one cluster.
Algorithms based on graph theory
The essence of such algorithms is that the sample of objects is represented as a graph G = (V, E) , whose vertices correspond to objects, and the edges have a weight equal to the “distance” between the objects. The advantages of graph clustering algorithms are visibility, relative ease of implementation, and the ability to make various improvements based on geometrical considerations. The main algorithms are the algorithm for selecting connected components, the algorithm for constructing the minimum spanning (spanning) tree and the layer-by-layer clustering algorithm.
Algorithm of selection of connected components
In the algorithm for selecting connected components, the input parameter R is specified and all edges for which “distances” are greater than R are removed in the graph. Only the closest pairs of objects remain connected. The meaning of the algorithm is to find a value of R that lies in the range of all “distances” at which the graph “falls apart” into several connected components. The resulting components are clusters.
For the selection of the parameter R , a histogram of distributions of pairwise distances is usually constructed. In problems with a well-pronounced cluster data structure, there will be two peaks in the histogram — one corresponds to intracluster distances, and the second to intercluster distances. The parameter R is selected from the minimum zone between these peaks. At the same time, managing the number of clusters using the distance threshold is rather difficult.
Algorithm minimum spanning tree
The minimum spanning tree algorithm first builds a minimal spanning tree on the graph, and then successively removes the edges with the greatest weight. The figure shows the minimum spanning tree obtained for nine objects.

By removing the link labeled CD, with a length of 6 units (edge with maximum distance), we obtain two clusters: {A, B, C} and {D, E, F, G, H, I}. The second cluster can be further divided into two more clusters by removing the edge EF, which has a length of 4.5 units.
Layered clustering
The layer-by-layer clustering algorithm is based on identifying connected components of a graph at some level of distance between objects (vertices). The distance level is set by the distance threshold c . For example, if the distance between objects
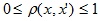 then
then 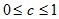 .
.The layer clustering algorithm generates a sequence of subgraphs of the graph G , which reflect the hierarchical relationships between clusters:
 ,
,where G t = (V, E t ) is a graph at the level of t ,
 ,
,with t - t-th threshold of distance,
m is the number of hierarchy levels,
G 0 = (V, o) , o is the empty set of edges of the graph obtained when t 0 = 1,
G m = G , that is, the graph of objects without restrictions on the distance (the length of the edges of the graph), since t m = 1.
By changing the distance thresholds { c 0 , ..., m }, where 0 = c 0 < c 1 <... < c m = 1, it is possible to control the depth of the hierarchy of the resulting clusters. Thus, a layer-by-layer clustering algorithm is able to create both flat data partitioning and hierarchical.
Algorithm Comparison
Computational complexity of algorithms
| Clustering algorithm | Computational complexity |
| Hierarchical | O (n 2 ) |
| k-medium | O (nkl), where k is the number of clusters, l is the number of iterations |
|---|---|
| c-medium | |
| Selection of connected components | depends on the algorithm |
| Minimum spanning tree | O (n 2 log n) |
| Layered clustering | O (max (n, m)), where m <n (n-1) / 2 |
Comparative table of algorithms
| Clustering algorithm | Cluster form | Input data | results |
| Hierarchical | Arbitrary | Number of clusters or distance threshold for truncating the hierarchy | Binary Cluster Tree |
| k-medium | Hypersphere | Number of clusters | Cluster Centers |
| c-medium | Hypersphere | The number of clusters, the degree of fuzziness | Cluster centers, membership matrix |
| Selection of connected components | Arbitrary | R threshold | Cluster tree structure |
| Minimum spanning tree | Arbitrary | Number of clusters or distance threshold for deleting edges | Cluster tree structure |
| Layered clustering | Arbitrary | Sequence of distance thresholds | Tree structure of clusters with different levels of hierarchy |
Little application
In my work, I needed to select separate areas from the hierarchical structures (trees). Those. in fact, it was necessary to cut the original tree into several smaller trees. Since an oriented tree is a special case of a graph, algorithms based on graph theory are naturally appropriate.
In contrast to a fully connected graph, in an oriented tree, not all vertices are connected by edges, and the total number of edges is n – 1, where n is the number of vertices. Those. applied to the nodes of the tree, the operation of the algorithm for selecting connected components will be simplified, since the removal of any number of edges will “decompose” the tree into connected components (individual trees). The algorithm of the minimum spanning tree in this case will coincide with the algorithm for selecting connected components — by removing the longest edges, the original tree is divided into several trees. It is obvious that the construction phase of the minimal spanning tree itself is skipped.
In the case of using other algorithms, they would have to separately consider the presence of connections between objects, which complicates the algorithm.
Separately, I want to say that in order to achieve the best result, it is necessary to experiment with the choice of distance measures, and sometimes even change the algorithm. There is no single solution.
Bibliography
1. Vorontsov K.V. Algorithms for clustering and multidimensional scaling . Lecture course. Moscow State University, 2007.
2. Jain A., Murty M., Flynn P. Data Clustering: A Review . // ACM Computing Surveys. 1999. Vol. 31, no. 3
3. Kotov A., Krasilnikov N. Data clustering . 2006
3. Mandel I. D. Cluster analysis. - M .: Finance and Statistics, 1988.
4. Applied statistics: classification and reduction of dimension. / S.A. Ayvazyan, V.M. Buchstaber, I.S. Enyukov, LD Meshalkin - M .: Finance and Statistics, 1989.
5. Information and analytical resource dedicated to machine learning, pattern recognition and data mining - www.machinelearning.ru
6. Chubukova I.A. Course of lectures "Data Mining", Internet University of Information Technologies - www.intuit.ru/department/database/datamining
Source: https://habr.com/ru/post/101338/
All Articles